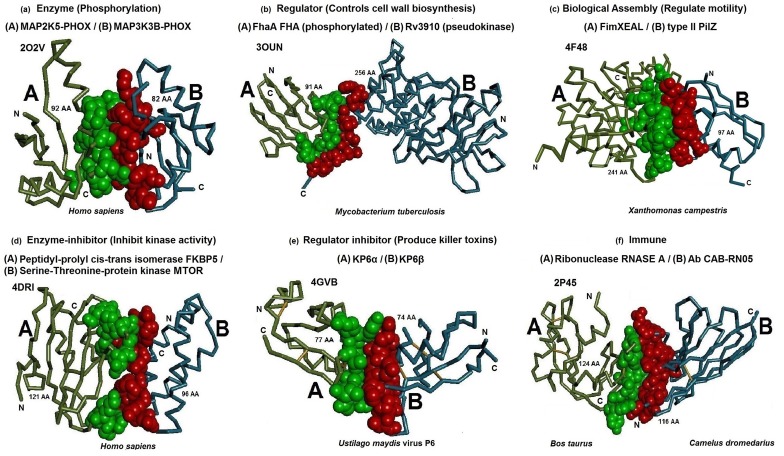
Figure 2.
Protein-protein complexes among different categories are shown using Discovery studio™ [28] with backbone structures displayed in Ca stick style and interface regions depicted using CPK (Corey Pauling Koltun) representation. The interface residues of chain A and B are colorized in green and red, respectively. (A) An enzyme complex (PDB ID 2O2V) between MAP2K5-PHOX / MAP3K3B-PHOX, (B) A regulator complex (PDB ID 3OUN) between FhaA FHA protein/Rv3910, (C) A protein assembly complex (PDB ID 4F48) formed between FimXEAL / type II PilZ, (D) An enzyme-inhibitor complex (PDB ID 4DRI) between Peptidyl-prolyl cistrans isomerase FKBP5 / Serine-Threonine-protein kinase MTOR, (E) A regulator inhibitor complex (PDB ID 4GVB) between KP6α/ KP6β and (F) An immune complex (PDB ID 2P45) between Ribonuclease RNASE A / Ab CAB-RN05. The interface residues were identified using change in Accessible Surface Area (ASA) [29] upon complex formation as described elsewhere [8, 13].