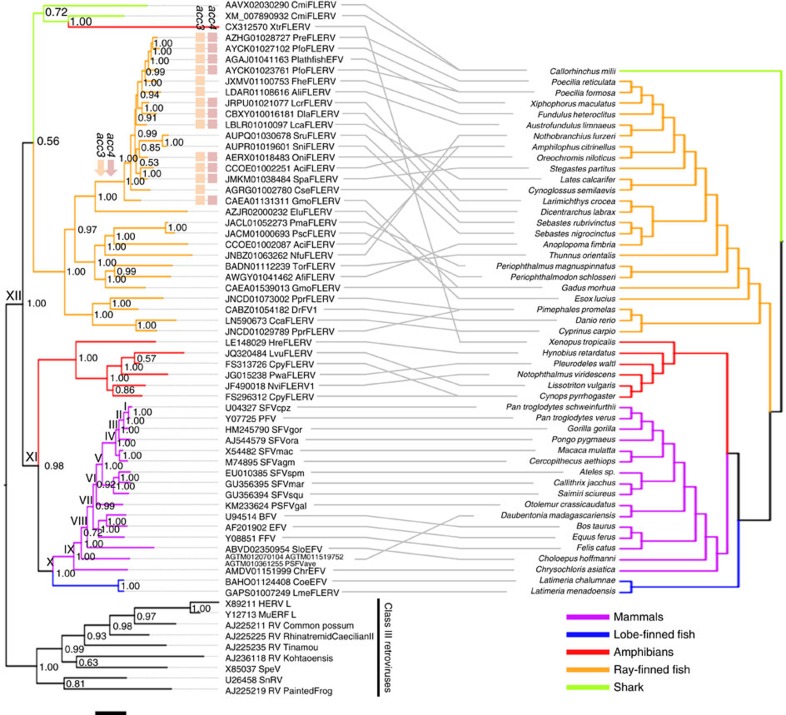
Figure 3. Coevolution of FVs and FLERVs and their vertebrate hosts.
A Bayesian phylogeny of FVs and FLERVs (left) is compared with the published vertebrate host cladogram35,36,37,38 (right). Preceding viral names are the contig accession numbers containing viral sequences. Class III retroviruses were used to root the viral tree. Solid grey lines between the two trees indicate viral–host associations. The scale bar (black solid line; underneath the virus phylogeny) represents genetic divergence of length 0.3 in units of amino acid substitutions per site, and Arabic numerals on nodes are Bayesian posterior probability node support values. Roman numerals indicate the nodes of which the total per-lineage substitutions to the chimpanzee simian FV (U14327 SFVcpz) were used to construct the time-dependent rate phenomenon model (Fig. 4). The presence of acc3 (orange squares) and acc4 (red squares) is mapped onto the viral phylogeny. The most parsimonious timing of acc3 and acc4 acquisition is indicated (orange and pink arrows, respectively).