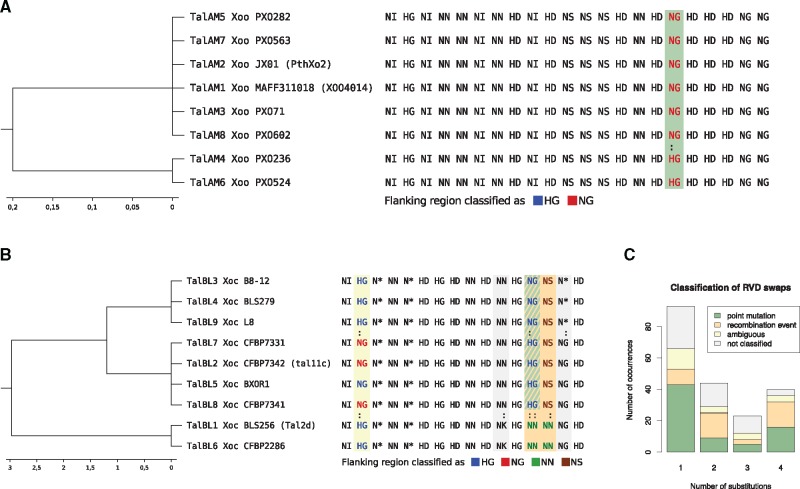
Fig. 3.
—Classification of RVD swaps by flanking sequence indicating point mutations or recombination. (A) Example for classification of flanking sequences in class TalAM. RVD swaps between different TALEs are marked with a colon. Common RVDs at swap positions are colored depending on the classification result of their flanking sequence by the corresponding binary classifier. In this case, all repeats at column 17 of the alignment are classified as NG repeats based on their flanking sequence, which indicates a point mutation in the RVD codon pair as indicated by the green shading of that column. (B) Example for classification of flanking sequences in class BL. RVD swaps between different TALEs are marked with a colon. Common RVDs at swap positions are colored depending on the classification result of their flanking sequence by the corresponding binary classifier. Columns with RVD swaps are shaded according to the colors used in C. (C) Results of classification for TALE repeats involved in RVD swaps in all AnnoTALE classes with four different outcomes: (a) all repeats are classified as the same RVD, which suggests point mutation within the RVD (green), (b) all flanking sequences are classified like the RVD of the respective RVD, indicating a recombination event (orange), (c) the classification is ambiguous (yellow). (d) No classifier trained for the combination of these two RVDs and repeats have not been classified (grey).