Abstract
Background
Bast fibres are characterized by very thick secondary cell walls containing high amounts of cellulose and low lignin contents in contrast to the heavily lignified cell walls typically found in the xylem tissues. To improve the quality of the fiber-based products in the future, a thorough understanding of the main cell wall polymer biosynthetic pathways is required. In this study we have carried out a characterization of the genes involved in lignin biosynthesis in flax along with some of their regulation mechanisms.
Results
We have first identified the members of the phenylpropanoid gene families through a combination of in silico approaches. The more specific lignin genes were further characterized by high throughput transcriptomic approaches in different organs and physiological conditions and their cell/tissue expression was localized in the stems, roots and leaves. Laccases play an important role in the polymerization of monolignols. This multigenic family was determined and a miRNA was identified to play a role in the posttranscriptional regulation by cleaving the transcripts of some specific genes shown to be expressed in lignified tissues. In situ hybridization also showed that the miRNA precursor was expressed in the young xylem cells located near the vascular cambium. The results obtained in this work also allowed us to determine that most of the genes involved in lignin biosynthesis are included in a unique co-expression cluster and that MYB transcription factors are potentially good candidates for regulating these genes.
Conclusions
Target engineering of cell walls to improve plant product quality requires good knowledge of the genes responsible for the production of the main polymers. For bast fiber plants such as flax, it is important to target the correct genes from the beginning since the difficulty to produce transgenic material does not make possible to test a large number of genes. Our work determined which of these genes could be potentially modified and showed that it was possible to target different regulatory pathways to modify lignification.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-1072-9) contains supplementary material, which is available to authorized users.
Keywords: Flax, Laccase, Lignin, microRNA, Monolignol, Phenylpropanoids, Stress
Background
Lignin is among the most abundant biological polymers on earth. It is a complex aromatic molecule synthesized during the onset of the secondary cell wall (SCW) formation in plants providing stiffness proprieties for mechanical strength, hydrophobicity for water transport and contributes to defense against pests and pathogens. Lignin is produced by a complex biosynthetic pathway (Fig. 1) which also leads to the production of a wide range of phenylpropanoids with diverse and sometimes unknown functions such as hydroxycinnamic acids, flavonoids, coumarins, chalcones, phenylpropenes and stilbenes [1]. In brief, phenylalanine derived from the shikimate pathway is used as an initial substrate [2]. This amino acid is first deaminated by phenylalanine ammonia-lyase (PAL; EC 4.3.1.5), hydroxylated by cinammate 4-hydroxylase (C4H; EC 1.14.13.11) and then esterified with CoA by 4-coumarate:CoA ligase (4CL; EC 6.2.1.12) forming p-coumaroyl CoA. This metabolite can lead to the formation of p-coumaryl alcohol by the action of cinnamoyl CoA reductase (CCR; EC 1.2.1.44) and cinnamyl alcohol dehydrogenase (CAD; EC 1.1.1.195). p-coumaroyl CoA can also be successively transformed by hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (HCT; EC 2.3.1.133) and p-coumarate 3-hydroxylase (C3’H; EC 1.14.14.1) to form caffeoyl CoA that can be methoxylated by caffeoyl CoA 3-O-methyltransferase (CCoAOMT; EC 2.1.1.104) into feruloyl CoA. This CoA ester can be reduced into coniferaldehyde by CCR and further transformed into coniferyl alcohol by CAD. Both molecules are hydroxylated by ferulate 5-hydroxylase (F5H; EC 1.14.13) and methoxylated by caffeate/5-hydroxyferulate O-methyl-transferase (COMT; EC 2.1.1.68) forming sinapaldehyde and sinapyl alcohols respectively. In addition to p-coumaryl alcohol, both alcohols are named monolignols. These can be transported to the apoplast and transformed into radicals by laccases and peroxidases [3, 4]. The oxidative coupling of these activated monolignols leads to the production of p-hydroxyphenyl (H), syringyl (S) and guaiacyl (G) units respectively, present in different proportions within the lignin fractions depending on the taxonomic families, species, tissues or environment [5]. The lignin biosynthesis pathway has been comprehensively characterized in model species such as Arabidopsis and the identified gene models then used to search for orthologous sequences in more economically-important plants including food crops as well as woody or fiber species.
Fig. 1.
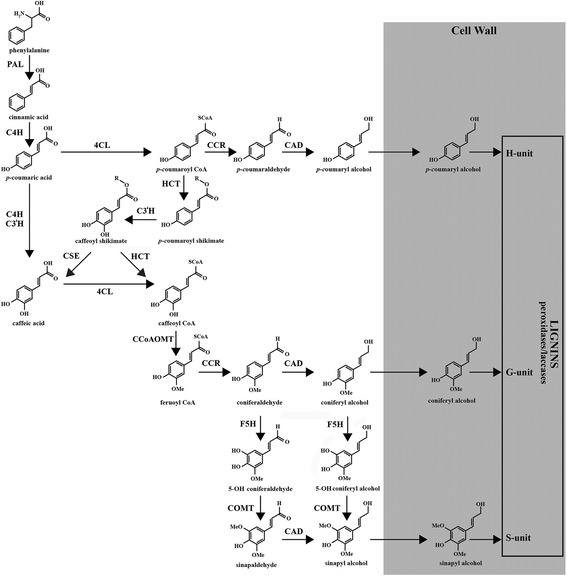
The monolignol and lignin biosynthetic pathway. 4CL: 4-coumarate:CoA ligase; BGLU: beta glucosidase; C3’H: p-coumarate 3-hydroxylase; C4H: cinnamate 4-hydroxylase; CAD: cinnamyl alcohol dehydrogenase; CCoAOMT: caffeoyl CoA 3-O-methyltransferase; CCR: cinnamoyl CoA reductase; COMT: caffeate/5-hydroxyferulate O-methyl-transferase; CSE: caffeoyl shikimate esterase; F5H: ferulate 5-hydroxylase; HCT: hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase; PAL: phenylalanine ammonia-lyase; UGT: UDP glycosyltransferase
Bast fibers are present in bundles within the stem cortex between the epidermis and the xylem core in some non-woody plants. These bundles may contain only primary fibers derived from the procambium in species such as flax (Linum usitatissimum) or ramie (Boehmeria nivea) while others such as jute (Corchorus species), kenaf (Hibiscus cannabinus) or hemp (Cannabis sativa) contain additional secondary fibers derived from the vascular cambium. Fiber species are considered as interesting biological models because their stems contain two main populations of cells showing highly contrasted SCW compositions [6]. The xylem cells from the inner-stem tissues have a typical SCW structure with up to 30% of the cell wall weight represented by lignins whereas the bast fibers possess cellulose-rich thick SCWs with lignin contents ranging from less than 1% (ramie) to 19% (kenaf) [7]. The cell wall structure and composition is very important in these species because they determine the properties of the extracted raw materials. Flax, for instance, has been used for several thousands of years to make ropes and linen tissues and more recently, for the production of environmentally friendly fiber-based composite materials [8, 9]. To optimize the use of these materials in the future, targeted engineering of the cell wall to improve product quality will require a thorough understanding of the biosynthetic pathways leading to the production of the main cell wall polymers. Over the past years, significant efforts including genome sequencing [10], development of molecular tools [11–13] and databases [14, 15] have allowed us not only to improve our understanding of flax fiber cell wall biology, but also to identify the different gene models potentially associated with the biosynthesis of different cell wall polymers. Nevertheless, many cell wall genes are part of multigenic families and a major challenge to engineering is the identification of the individual family members involved in lignification. For non-model plants such as flax, it is important to target the correct genes from the beginning since rapid and easy transformation to obtain high amounts of transgenic material is particularly difficult [16].
To clearly identify the genes that are indeed involved in flax lignin production, we first conducted a high throughput reverse-transcriptase quantitative PCR (HT-RT-qPCR) approach to establish the expression profile of in silico-identified genes using organs/tissues with contrasted lignin contents. To further characterize these genes and because it was not possible to use large-scale reporter gene transformation, we used in situ hybridization to confirm the expression of the identified genes in lignified tissues. This type of approach has already been used to detect some lignin gene transcripts in woody plants [17–19] but never on such a large scale. We also benefited from the development of this technique in flax to show that a microRNA, miR397, already shown to be involved in the cleavage of several laccase transcripts [20, 21] is co-expressed in the same cells as the lignin biosynthetic genes confirming the role of miRNAs on laccase regulation in flax. In the context of a possible impact of global climate changes on the quality of plant products, and because lignin is an important component in stress responses [5], the expression of the identified genes was also analysed under a range of different environmental conditions. The high amount of data obtained by HT-RT-qPCR has enabled us to show that the expression of most of these genes is coordinated, with a probable implication of MYB transcription factors.
Results
Identification of genes related to phenylpropanoid biosynthesis in flax
The whole genome shotgun assembly of flax [10] was first surveyed to identify the genes potentially related to phenylpropanoid biosynthesis. BLAST analyses were carried out to search against the 88,420 scaffolds including 43,471 gene models. A total of 69 genes were identified and organized in gene families containing between 3 and 15 members (Table 1) and the presence of the corresponding transcripts for each gene was checked among the different public EST databases. Only Lus4CL8, Lus4CL9 and LusCCoAOMT5 have no corresponding ESTs (E value <e-50) and the expression of LusHCT1, LusCCR3, LusCCR8, LusCAD8 and LusCAD11 was also undetectable when using whole genome microarrays [22]. In addition to these 8 genes, no expression data were obtained for LusCCR9, LusCCR11, LusCCR12 and LusCAD9 using EST-based microarrays [11, 23]. The intron/exon structure of the genes is graphically represented in Additional file 1: Figure S1.
Table 1.
Characteristics of the flax phenylpropanoid genes identified in this work
| Gene name | Gene model | Genome | Microarray EST | GenBank EST | Ref |
|---|---|---|---|---|---|
| LusPAL1 | Lus10040416 | scaffold86 | c247 | JG065006 | 5 |
| LusPAL2 | Lus10023531 | scaffold1216 | c247 | EH791565 | 4 |
| LusPAL3 | Lus10026518 | scaffold617 | c904 | JG216766 | 5 |
| LusPAL4 | Lus10013805 | scaffold618 | c904 | JG216766 | 5 |
| LusC4H1 | Lus10034449 | scaffold310 | c1220 | JG081734 | 5 |
| LusC4H2 | Lus10019110 | scaffold30 | c32549 | JG085936 | 1;5 |
| LusC4H3 | Lus10021671 | scaffold208 | c6570 | GW864597 | 2 |
| LusC4H4 | Lus10035011 | scaffold43 | c4373 | JG075414 | 1;5 |
| LusC4H5 | Lus10027598 | scaffold2 | c11079 | ||
| Lus4CL1 | Lus10026143 | scaffold319 | c3640 | CA483243 | |
| Lus4CL2 | Lus10008677 | scaffold1635 | c3640 | CA483243 | |
| Lus4CL3 | Lus10024123 | scaffold353 | c680 | JG220816 | 5 |
| Lus4CL4 | Lus10005390 | scaffold547 | c680 | JG218659 | 5 |
| Lus4CL5 | Lus10002791 | scaffold125 | c15833 | JG239276 | 5 |
| Lus4CL6 | Lus10016135 | scaffold344 | c3980 | EH791222 | 4 |
| Lus4CL7 | Lus10021431 | scaffold612 | c3980 | GW866203 | 2 |
| Lus4CL8 | Lus10025842 | scaffold605 | |||
| Lus4CL9 | Lus10038259 | scaffold28 | |||
| LusHCT1 | Lus10002321 | scaffold120 | JG213400 | 5 | |
| LusHCT2 | Lus10026097 | scaffold319 | c3481 | JG235206 | 5 |
| LusHCT3 | Lus10010786 | scaffold18 | c6233 | JG202486 | 5 |
| LusHCT4 | Lus10026123 | scaffold319 | c8096 | CV478884 | 2 |
| LusHCT5 | Lus10022163 | scaffold342 | c6233 | JG202486 | 5 |
| LusC3’H1 | Lus10033524 | scaffold701 | c581 | JG230193 | 5 |
| LusC3’H2 | Lus10020847 | scaffold711 | c5820 | JG226878 | 5 |
| LusC3’H3 | Lus10020850 | scaffold711 | c5820 | CA483318 | |
| LusCCoAOMT1 | Lus10027888 | scaffold1143 | c395 | JG228418 | 5 |
| LusCCoAOMT2 | Lus10002837 | scaffold810 | c395 | JG228418 | 5 |
| LusCCoAOMT3 | Lus10019841 | scaffold1491 | c2503 | JG094269 | 5 |
| LusCCoAOMT4 | Lus10014074 | scaffold1247 | c2503 | JG231505 | 3;5 |
| LusCCoAOMT5 | Lus10034584 | scaffold9 | |||
| LusCCR1 | Lus10041651 | scaffold272 | c5445 | JG227337 | 5 |
| LusCCR2 | Lus10024068 | scaffold353 | c2262 | GW866594 | 2 |
| LusCCR3 | Lus10008774 | scaffold729 | JG203738 | 5 | |
| LusCCR4 | Lus10022239 | scaffold225 | c11788 | GW865040 | 2 |
| LusCCR5 | Lus10024138 | scaffold353 | c13273 | JG149727 | 5 |
| LusCCR6 | Lus10042399 | scaffold123 | c20639 | JG103050 | 5 |
| LusCCR7 | Lus10026273 | scaffold898 | c20639 | JG103050 | 5 |
| LusCCR8 | Lus10006885 | scaffold329 | JG097906 | 5 | |
| LusCCR9 | Lus10003780 | scaffold806 | JG088940 | 5 | |
| LusCCR10 | Lus10012930 | scaffold434 | c6761 | JG215733 | 5 |
| LusCCR11 | Lus10030973 | scaffold261 | JG240727 | 5 | |
| LusCCR12 | Lus10035369 | scaffold151 | JG240727 | 5 | |
| LusF5H1 | Lus10028361 | scaffold413 | c772 | JG214534 | 5 |
| LusF5H2 | Lus10041811 | scaffold272 | c772 | JG214534 | 5 |
| LusF5H3 | Lus10014273 | scaffold275 | c2179 | GW865908 | 2 |
| LusF5H4 | Lus10025975 | scaffold319 | c2179 | GW867449 | 2 |
| LusF5H5 | Lus10034300 | scaffold310 | c56836 | ||
| LusF5H6 | Lus10012582 | scaffold6 | c56836 | ||
| LusF5H7 | Lus10022303 | scaffold225 | c56836 | ||
| LusF5H8 | Lus10041511 | scaffold272 | c56836 | ||
| LusCOMT1 | Lus10015576 | scaffold233 | c2253 | JG229091 | 5 |
| LusCOMT2 | Lus10032929 | scaffold51 | c629 | JG229091 | 5 |
| LusCOMT3 | Lus10009442 | scaffold981 | c4476 | GW865137 | 2 |
| LusCAD1 | Lus10027864 | scaffold1143 | c2456 | JG020400 | 5 |
| LusCAD2 | Lus10002812 | scaffold810 | c2456 | JG020400 | 5 |
| LusCAD3 | Lus10019811 | scaffold1491 | c3049 | GW865020 | 2 |
| LusCAD4 | Lus10014104 | scaffold1247 | c3049 | JG215109 | 5;6 |
| LusCAD5 | Lus10014363 | scaffold275 | c4852 | JG019640 | 5 |
| LusCAD6 | Lus10010149 | scaffold587 | c6705 | GW867690 | 2 |
| LusCAD7 | Lus10026070 | scaffold319 | c4852 | JG102096 | 5 |
| LusCAD8 | Lus10023268 | scaffold98 | JG062652 | 5 | |
| LusCAD9 | Lus10035956 | scaffold76 | EH792205 | 4 | |
| LusCAD10 | Lus10002089 | scaffold575 | c4394 | JG255907 | 5 |
| LusCAD11 | Lus10025706 | scaffold605 | GW866334 | 2 | |
| LusCAD12 | Lus10002302 | scaffold120 | c5057 | JG282203 | 5 |
| LusCAD13 | Lus10002300 | scaffold120 | c6631 | JG216469 | 5 |
| LusCAD14 | Lus10009955 | scaffold200 | c3693 | JG140771 | 5 |
| LusCAD15 | Lus10039595 | scaffold15 | c3758 | JG217534 | 5 |
1: PR Babu, KV Rao and VD Reddy [86]); 2: A day, M Addi, W Kim, H David, F Bert, P Mesnage, C Rolando, B Chabbert, G Neutelings and S Hawkins [87]); 3: A day, G Neutelings, F Nolin, S Grec, a Habrant, D Cronier, B Maher, C Rolando, H David, B Chabbert, et al. [30]); 4: MJ roach and MK Deyholos [23]); 5: P Venglat, D Xiang, S Qiu, SL stone, C Tibiche, D cram, M Alting-Mees, J Nowak, S Cloutier, M Deyholos, et al. [88]); 6: M Wrobel-Kwiatkowska, M Starzycki, J Zebrowski, J Oszmianski and J Szopa [60])
In silico and expression approaches to identify monolignol biosynthetic genes
To identify the genes implicated in lignin precursor biosynthesis, a phylogenetic analysis was first performed by combining the protein sequences derived from the flax phenylpropanoid genes with those from Arabidopsis thaliana, Vitis vinifera and Populus trichocarpa (Additional file 2: Figure S2). Different bona fide proteins characterized at the biochemical level or by forward/reverse genetic approaches previously used to identify lignin genes in Eucalyptus grandis [24] were also added to the data. In addition to this in silico sequence comparison, we also performed HT-RT-qPCR on whole stems, roots and leaves as well as on inner stem xylem-rich tissues and outer stem bast fiber-rich tissues (Fig. 2). Gene expression was also determined in leaves and whole stems under different stress conditions (Additional file 3: Figure S3). Due to the high conservation of several gene sequences and intron/exon positions in some clades, but also the lack of annotation of the more specific 5′/3′-UTRs portions in the genome, we sometimes had to design primers targeting several close-related genes. Taken together, we performed amplifications on 35 single, 12 double, 2 triple and 4 quadruple gene groups (Additional file 4: Table S1). In our experimental conditions, we were unable to detect the expression of LusPAL4, LusC4H4, Lus4CL3, Lus4CL6, LusCCR10, LusCCR12, LusF5H5, LusF5H6, LusF5H7, LusF5H8 and LusCAD6 genes whatever the organs or environmental conditions. We then used both the phylogenetic analyses (Additional file 2: Figure S2) and the HT-RT-qPCR (Fig. 2) data to identify the potential lignin-specific genes in flax. The major conclusions are summarized in the following subchapters.
Fig. 2.
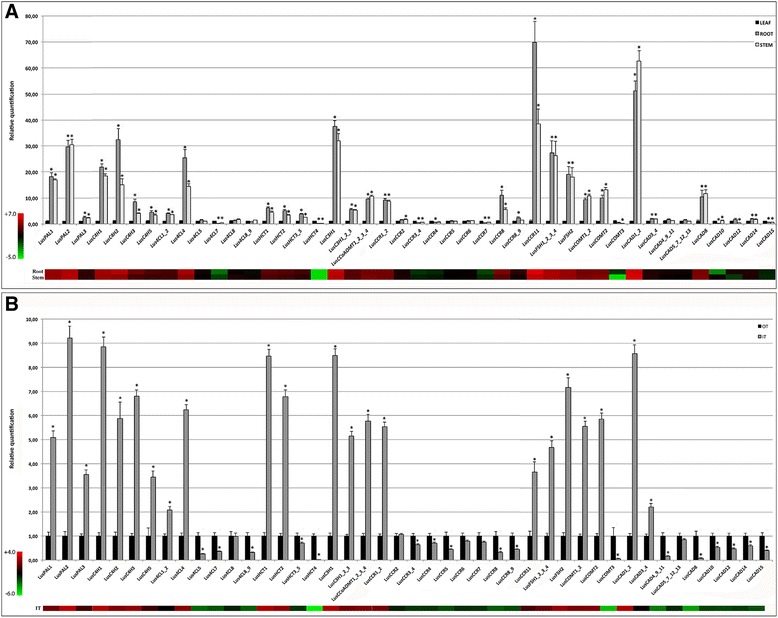
Expression of the flax phenylpropanoid genes determined by HT-RT-qPCR. a gene expression in leaves, roots and whole stems. b gene expression in the inner xylem-rich tissues (IT) and outer fiber-rich stem tissues (OT) of the stem. Values are means ± SD (n = 9). Asterisks indicates values that were determined significantly different from their control (a: Leaf and b: OT) using a Mann & Whitney’s test (* P < 0.01). The heat maps under the histograms represent the expression values in the roots and stems compared to the leaves (a) or in the IT compared to the OT (b). The colour codes and relative expression ranges in log2 values are represented on the left. * = significant difference in expression
PAL
The deamination step catalysed by the PAL enzymes leads to the formation of cinnamic acid, which may further undergo a series of esterification, hydroxylation and methylation steps. There are no clear reports on a single “lignin-specific” PAL gene in the usual plant models such as Arabidopsis or Populus because of a probable functional redundancy between the members of this gene family. For this reason, it was not easy to highlight PAL genes that are essential for lignin biosynthesis in flax when using only a phylogenetic approach. The 4 flax PAL genes identified in this work were all located among the bona fide genes. Although the bootstrap values were sometimes low among the subdivisions in this part of the tree, LusPAL1 and LusPAL2 were closely related to the 3 xylem-specific poplar genes PtrPAL2, PtrPAL4 and PtrPAL5 [25] while LusPAL3 and LusPAL4 were closer to the Arabidopsis orthologs AthPAL1 and AthPAL2 involved in lignin metabolism [26]. LusPAL1 and LusPAL2 are highly expressed in stems and roots compared to leaves. Their respective expression levels in the inner stem (xylem tissues) are close to five- and nine-fold higher than that observed in the hypolignified external stem tissues. Consequently, LusPAL1 and LusPAL2 are the most likely candidates for lignin biosynthesis in flax.
C4H
C4H (CYP73) is the first of the three cytochrome P450 monooxygenases involved in the hydroxylation of phenylpropanoid precursors. In flax, five distinct genes located on different scaffolds were annotated by KEGG as trans-cinnamate 4-monooxygenases (K00487). In the neighbour joining tree, 2 distinct clades were present, one contained all of the bona fide functionally characterized orthologs, together with an additional Vitis sequence and the flax proteins LusC4H1–4. In the other clade, LusC4H5 was located with PtrC4H3 and the eucalyptus EgrC4H1, suggested to be the main C4H involved in lignin biosynthesis [24] even though EgrC4H2 was also preferentially expressed in the xylem. The HT-RT-qPCR profiles of the flax genes showed that LusC4H1, LusC4H2, LusC4H3 and LusC4H5 were expressed in lignified tissues with highest levels for LusC4H1–2 making them the best candidates for a major role in lignification.
4CL
The 4CL enzymes produce specific CoA thioesters of 4-hydroxycinnamic acids at an important crossroad in the phenylpropanoid pathway. Nine 4CL gene models were identified in the genome of Linum usitatissimum. When the major known sequences were assembled in the phylogenetic tree, a delimited bona fide clade containing two separated classes was identified as shown in previous studies [27, 28]. The class I, predicted to be involved in monolignol biosynthesis, contained four flax proteins Lus4CL1–4. The Lus4CL3 gene remained undetectable in our conditions whereas the other three genes had higher expression in roots and in stems. The relative expression of Lus4CL4 was 6 fold higher in the xylem as compared to the hypolignified external stem tissues and 15- and 25- fold higher in the stems and roots as compared to the leaves and so is likely involved in lignin biosynthesis.
HCT
In the lignin biosynthetic pathway, HCTs first catalyse the formation of p-coumaroyl shikimate from p-coumaroyl CoA and shikimic acid. To produce caffeoyl CoA, the product is first hydroxylated by C3’H and then converted again by HCT acting in the reverse direction. The closely related acyltransferase hydroxyl-cinnamoyl CoA:quinate hydroxycinnamoyl transferase (HQT) can lead to the formation of chlorogenic acid by the transfer of quinic acid on the same p-coumaroyl CoA substrate, forming p-coumaroylquinic acid. The distinction between HCT and HQT genes without further biochemical characterization is not always very easy because of their very similar sequences. We identified five different genes in the flax genome closely related to the previous identified HCT/HQT genes. The expression profiles show that LusHCT1 and LusHCT2 are active in the roots and highly expressed in stem-internal tissues. Their position among the bona fide HCT sequences in the phylogenetic tree also suggests their implication in lignin biosynthesis. The LusHCT4 gene is also among the true HCTs but the expression of this gene is approximately 50 fold times higher in the hypolignified external tissues of the stem.
C3’H
The C3’H genes generally belong to small hydroxylase subfamilies involved in monolignol biosynthesis. The enzyme (CYP98A3) catalyses the 3′-hydroxylation of 4-coumaroyl shikimate and 4-coumaroyl quinate into the corresponding caffeoyl-conjugated form. In flax, we identified only 3 gene models corresponding to C3’H. A gene duplication event possibly occurred because LusC3’H2 and LusC3’H3 are both located on the same scaffold in a 14 kb region. When considering the structure (Additional file 1: Figure S1) and homology between the 3 genes, it is possible to speculate that a first duplication event of LusC3’H1 formed the LusC3’H2 gene, which in turn was duplicated forming LusC3’H3. This gene was shorter at the 3′-end but still had between 92 and 98% amino acid sequence identity with the 2 former corresponding proteins. In Arabidopsis only one C3’H gene was identified whereas an expansion of this family by lineage-specific tandem duplications in Populus [29] and Eucalyptus [24] was described. The LusC3’H1 gene is more strongly expressed in the stem and root tissues and therefore probably involved in lignin biosynthesis.
CCoAOMT
Among both methylation enzyme families, CCoAOMTs can transfer a –CH3 group from a donor to a hydroxycinnamoyl CoA ester, namely caffeoyl CoA. Among the 5 genes identified in this work, LusCCoAOMT4 was previously characterized in knockdown flax plants [30]. The four genes LusCCoAOMT1–4 have high sequence identity, similar gene structures and could not be individualized by the HT-RT-qPCR approach. The amplicon was detected at high levels in lignin-containing organs such as stems and roots compared to leaves and preferentially in the internal stem tissues. In the phylogenetic tree, they collocate with the sequences identified as bona fide enzymes in Arabidopsis, Eucalyptus, Nicotiana and Populus.
CCR
This enzyme is important because it catalyses the first specific step of the monolignol production by reducing hydroxycinnamoyl CoA esters. Taking into account the large size of this gene family as determined by genome sequencing, they are likely to be involved in numerous other metabolic pathways. In flax, we identified 12 genes with very different intron/exon patterns. The LusCCR3, LusCCR5 and LusCCR6 genes do not fit the 3 patterns described for Arabidopsis [31]. Although most of the genes encode proteins with similar sizes, LusCCR3 appears smaller and thus may be truncated. In Arabidopsis, 12 CCR genes were present in duplicated chromosome regions including 4 genes distributed in tandem [31]. No similar situation exists in flax although LusCCR2 and LusCCR5 were located on the same scaffold but at a distance of 360 kb. The differences in the structure of these 2 genes are however not in favour of a recent duplication event. The bona fide lignin biosynthesis genes were grouped in a common clade also containing LusCCR1, LusCCR2, LusCCR11 and LusCCR12. The expression profiles of these four genes in the lignified tissues showed that LusCCR1 and LusCCR11 are the best lignin-associated candidates.
COMT
As indicated by the enzyme name, the caffeate/5 hydroxyferulate O-methyl-transferase was first thought to act as a bifunctional enzyme able to transfer a methyl group from S-adenosyl methionine to the C3 and C5 positions of the aromatic rings of caffeic and 5-hydroxyferulic acids [32]. In fact, the COMT enzyme promotes the biochemical pathway leading to the formation of sinapyl alcohol by catalysing the methylation of 5-hydroxyconiferyl alcohol [33]. Only 3 gene models were identified within the flax COMT gene family, which is low compared to many other species [34]. LusCOMT1 and LusCOMT2 have close exon/intron patterns but are present on different scaffolds. They both are expressed at higher rates in lignified tissues. The position of the corresponding proteins within the bona fide group in the phylogenetic tree seems to support the hypothesis of their implication in the lignin biosynthetic pathway.
F5H
The F5H cytochrome P450-dependent monooxygenase is required for the hydroxylation of coniferaldehyde and coniferyl alcohol leading to the formation of S lignin through the synthesis of sinapyl alcohol [35]. In flax, 8 gene models were identified and compared to orthologous models expressed in lignin-rich tissues. In the phylogenetic tree, these proteins are located on a branch separated from the bona fide enzymes by a node predicted by a low bootstrap value. The flax proteins were further subdivided into 2 groups of sequences with similar exon/intron patterns. The first group contained F5H1, F5H2, F5H3 and F5H4, which could not clearly be separated by HT-RT-qPCR during the design of the primers except for F5H2. The common F5H1_2_3_4 and specific F5H2 amplicons were detected at high levels in lignin-containing organs such as roots and whole stems and also in xylem-rich outer tissues. The second group with F5H5, F5H6, F5H7 and F5H8 genes had undetectable expression.
CAD
The NADPH-dependent CAD enzymes play an important role in monolignol biosynthesis by catalyzing the reduction of hydroxycinnamyl aldehydes into their corresponding monolignols which is the last step in the biosynthetic pathway [36]. They are encoded by a multigenic family and their homologs have been detected widely in bacteria and eukaryota but not in animals [37]. Classification by several authors highlighted 3 to 7 groups/classes based on phylogenetic approaches [38–41]. Recently an accurate classification proposed a separation in 3 functional clades for both dicots and monocots [42] with a lignin specific class I and a possible lignin-associated class II. In the present study we identified 15 flax gene models predicted in the published genome. Both LusCAD12 and LusCAD13 were located on scaffold120 in a 5.5 kb fragment and have the same exon/intron pattern associated with a high (84%) amino acid sequence identity, reflecting a possible duplication event. LusCAD1, LusCAD2, LusCAD3 and LusCAD4 segregate with the known bona fide genes within the phylogenetic tree. Because of their very high sequence identity (98,6% at the protein level), it was not possible to separate LusCAD1 and LusCAD2 when designing the primers for qRT-PCR. Their expression profiles show that the amplicons were mostly present in the lignified tissues such as roots and whole stems and also in xylem-rich outer tissues. To a lesser extent, LusCAD8 showed the same expression profile in these organs but revealed an opposite behaviour when comparing outer and inner stem tissues. This gene is located outside the bona fide clade.
Cell−/tissue-specific expression of lignin and selected lignin biosynthetic genes
The likely implication of identified candidate genes in the flax lignification process was further confirmed using in situ hybridization that established a close spatial correlation between lignifying cells determined by the Wiesner reaction and gene expression in stems, roots and leaves (Fig. 3 and summarized in Additional file 5: Table S2). These genes were LusPAL1, LusC4H2, Lus4CL4, LusHCT1, LusC3’H1_2_3, LusCCoAOMT1_2_3_4, LusCCR1_2, LusF5H1_2_3_4, LusCOMT1_2 and LusCAD1_2. For most genes, the strongest expression was observed in the stems and the roots in the first 2–5 secondary xylem cell layers from the cambial zone towards the inner part of the organs. In the leaves, the stain was in each case restricted to the primary xylem. For all genes, signal intensity was generally higher in stems and roots compared to leaves except for CCR for which equal intensity was observed in all 3 organs. In addition, the 4CL4 probe failed to reveal the presence of the corresponding transcripts in the stems and roots despite the clear organ specificity revealed by the HT-RT-qPCR results. The observed lack of signal might be due to poor probe efficiency since several fragments of the 4CL4 gene were tested but did not produce a positive signal in roots or stems. Overall, these results confirm that the expression of the selected genes is intimately associated with the actively differentiating secondary xylem zone thereby confirming their probable implication in the lignification process.
Fig. 3.
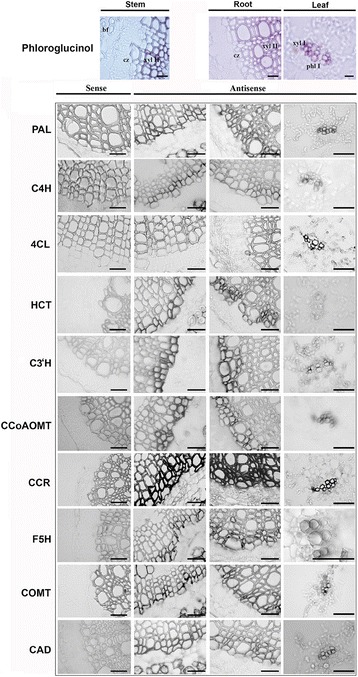
Lignin and lignin gene transcript localization by in situ hybridization in flax stems, roots and leaves. A set of paraffin stem sections (10 μm-thick) were first hybridized with the corresponding sense probes (left column) in order to confirm the specificity of the signal. Bar = 50 μm. The top panels show lignin localization (in red) by phloroglucinol-HCl staining. Phl I: primary phloem; xyl I: primary xylem; xyl II: secondary xylem; cz: cambial zone; bf: bast fiber. Bar = 50 μm
Identification of laccase genes and their regulation by microRNAs
Laccases play an important role in controlling lignin polymerization by catalysing the oxidation of the precursors [43]. Both in silico prediction and high throughput sequencing in different species [20, 21] have shown that laccase transcript levels are regulated by microRNAs. We identified 45 corresponding gene models in flax and used them to construct a phylogenetic tree (Fig. 4a). In comparison to previous published data reporting the classification of laccases in 6 clades [21], our tree contained an additional clade with 7 flax sequences. The expression profiles (Fig. 4b) of these laccase genes were extracted from public high throughput transcriptomic data [11, 15]. Of the 45 genes, 21 showed higher expression in ‘high lignin’ inner tissues compared to ‘low lignin’ outer tissues and are preferentially expressed on the top part of the stem where the development of the SCW takes place.
Fig. 4.
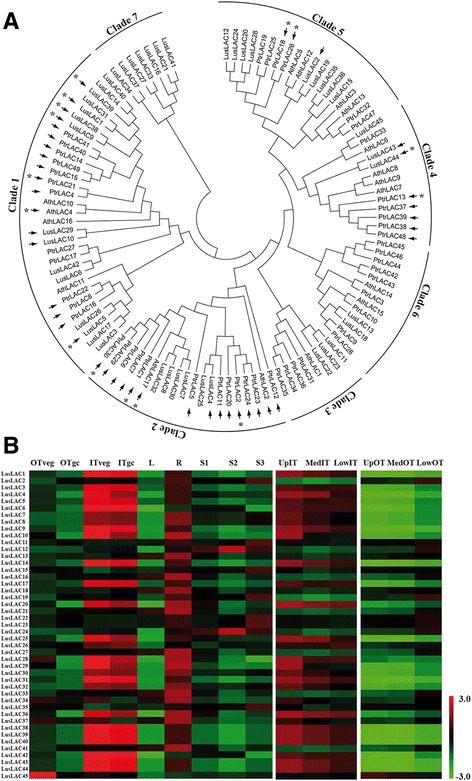
Laccase gene family in flax. a A phylogenetic tree was produced in MEGA 6 using the maximum likelihood method based on the JTT matrix-based model. The analysis was performed with 45 flax, 17 Arabidopsis, and 49 poplar laccase sequences. The arrows indicate the targets predicted by psRNAtarget and the asterisks, the experimentally validated targets. b Heat map showing laccase gene expression in different flax organs and tissues
Based on previous studies reporting predictive and experimental data on the cleavage of LAC4 and LAC17 transcripts in Arabidopsis [20] and potentially 29 laccase transcripts in Populus [21] by miR397, we first searched for a flax orthologous miRNA precursor sequence in the EST and genome databases as previously described [44]. The c3244 EST from the genolin database [11] and scaffold1999 extracted from the phytozome genomic database [45] both contained the pre-miRNA sequence (Fig. 5). The deduced mature lus-miR397 sequence was first used to search for targets among the flax laccases and among these, 11 genes were predicted with penalty mismatches included between 0 and 2.5 within the duplex. The amplification and sequencing of the 5′-end of the degraded transcripts by RLM-RACE allowed us to experimentally validate the cleavage of LusLAC1, LusLAC5, LusLAC9, LusLAC38, LusLAC39 and LusLAC44 among the predicted targets (Fig. 5). These 6 genes were present among the 21 genes highly expressed in the xylem-rich tissues. The five first laccase transcripts are close to the Arabidopsis lignin-related AthLAC4 gene [43] and all belong to the clade 1 (Fig. 4A). LusLAC44 belongs to clade 4 including PtrLAC13 which was shown to be cleaved by Ptr-miR397 [46]. These results demonstrate that flax laccase genes are regulated by microRNAs suggesting that this mechanism is implicated in controlling lignification in this species. Further evidence supporting this idea was provided by in situ hybridisation data (Fig. 6) indicating a close correlation between tissue−/cell-specific expression of the lus-miR397 precursor, monolignol specific genes and lignified tissues in stems and roots.
Fig. 5.
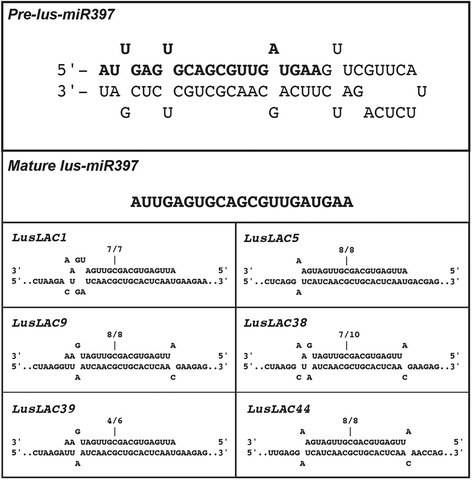
Lus-miR397 structure and validation of predicted targets. The sequences of the pre-lus-miR397 and the mature lus-miR397 are indicated. The association with laccase targets are shown only for the six transcripts validated by RLM 5′-RACE. The cleavage sites are indicated above the corresponding position and the number of cloned RACE products sequenced is shown above each sequence (number of sequenced fragments at this position/total number of sequenced clones)
Fig. 6.
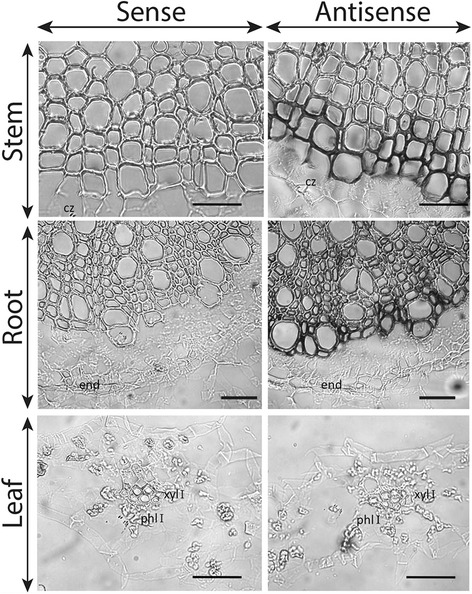
Spatial localization of lus-mir397 transcripts in flax organs. Paraffin sections (10 μm) of the stems, roots and leaves were hybridized with the sense controls (left) and the antisense specific probes (right). cz: cambial zone; end: endodermis; phl I: primary phloem; xyl I: primary xylem. Bar = 20 μm
Stress-related flax phenylpropanoid gene expression
Since lignin is an important component in stress responses, we analysed the expression of the phenylpropanoid genes with a special focus on the previous identified lignin-associated genes to search for those regulated by environmental modifications. Flax plants and isolated organs were submitted to a range of different stress conditions and gene expression determined by HT-RT-qPCR. The expression profiles were shown in Additional file 3: Fig. S3 and summarized in Table 2. Salicylic acid (SA) plays an important role in plant defense signaling and accumulates in response to pathogen infection [47]. In flax stems, SA significantly down-regulated the lignin-related genes LusC4H1 and LusCCoAOMT1_2_3_4 but had an opposite effect on LusPAL2 and the LusF5H1_2_3_4 group. Higher expression levels were also observed for Lus4CL8 and LusCAD10. In parallel, SA had a moderate effect on the expression of lignin genes in treated leaves. Only the up-regulation of the LusCAD4_9_11 group and LusCAD8 were evident. Methyljasmonate is a precursor of the active jasmonate form in plants [48] and mimics biotic as well as abiotic stresses [49]. MeJA application led to a very strong down-regulation of most of the phenylpropanoid genes including all of the identified bona fide lignin genes within the leaves except for LusCCR11. On the contrary, this signal regulator has a low overall effect on gene expression in flax stems. It is also interesting to note that some genes show no detectable expression levels in either the presence or absence of SA and MeJA, possibly due to the in vitro conditions used for organ incubation.
Table 2.
Summary of the stress response of flax phenylpropanoid genes
| Gene | Stress | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SA | MeJA | Drought | Wound | Light | |||||
| Stem | Leaf | Stem | Leaf | Stem | Leaf | Stem | Leaf | Stem | |
| LusPAL1* | − | − | + | − | |||||
| LusPAL2* | + | − | − | + | − | ||||
| LusPAL3 | − | − | − | + | |||||
| LusPAL4 | |||||||||
| LusC4H1* | − | − | − | + | + | + | |||
| LusC4H2* | − | + | + | ||||||
| LusC4H3 | − | + | + | ||||||
| LusC4H4 | |||||||||
| LusC4H5 | − | ||||||||
| Lus4CL1_2 | − | + | + | + | |||||
| Lus4CL3 | |||||||||
| Lus4CL4* | − | − | + | − | |||||
| Lus4CL5 | + | − | − | − | + | + | + | + | |
| Lus4CL6 | |||||||||
| Lus4CL7 | − | − | − | ||||||
| Lus4CL8 | + | − | |||||||
| Lus4CL8_9 | + | − | − | + | |||||
| LusHCT1* | − | − | + | ||||||
| LusHCT2* | − | − | + | − | |||||
| LusHCT3_5 | + | − | − | + | + | + | + | + | |
| LusHCT4 | − | − | − | − | |||||
| LusHCT5 | |||||||||
| LusC3’H1* | − | − | + | + | |||||
| LusC3’H1_2_3 | − | − | + | ||||||
| LusCCoAOMT1_2_3_4* | − | − | − | ||||||
| LusCCoAOMT5 | |||||||||
| LusCCR1_2* | − | − | |||||||
| LusCCR2 | − | − | + | ||||||
| LusCCR3_4 | + | ||||||||
| LusCCR4 | + | ||||||||
| LusCCR5 | + | − | − | + | + | + | + | ||
| LusCCR6 | − | − | |||||||
| LusCCR7 | − | − | |||||||
| LusCCR8 | − | ||||||||
| LusCCR8_9 | − | + | + | ||||||
| LusCCR10 | |||||||||
| LusCCR11* | + | + | + | ||||||
| LusCCR12 | |||||||||
| LusF5H1_2_3_4* | + | + | − | + | + | + | |||
| LusF5H2 | + | − | − | + | + | + | |||
| LusF5H5 | |||||||||
| LusF5H6 | |||||||||
| LusF5H7 | |||||||||
| LusF5H8 | |||||||||
| LusCOMT1_2* | − | + | + | + | |||||
| LusCOMT2* | + | − | + | ||||||
| LusCOMT3 | − | − | + | ||||||
| LusCAD1_2* | − | − | + | + | |||||
| LusCAD3_4 | + | − | − | + | + | ||||
| LusCAD4_9_11 | − | + | − | + | + | + | |||
| LusCAD5_7_12_13 | − | + | |||||||
| LusCAD6 | |||||||||
| LusCAD8 | − | + | − | + | + | + | |||
| LusCAD10 | + | − | + | + | + | ||||
| LusCAD12 | − | − | + | + | + | ||||
| LusCAD14 | + | + | + | + | |||||
| LusCAD15 | + | − | + | + | + | ||||
Asterisks indicate potential lignin-specific flax genes
The effect of abiotic stresses was further tested by inducing water deficit on whole flax plants grown in soil. Under these conditions, LusC4H1 and LusCCR11 were the only lignin-associated genes showing higher expression in leaves while most of the other bona fide genes were slightly upregulated in the stems. The effect of wounding was also tested on flax stems and leaves. In these organs, the effect on the lignin-associated genes was much stronger in the stems compared to the leaves. The modulation of phenylpropanoid gene expression was also evaluated after 48 h of continuous light illumination. When compared to 16 h/8 h light/dark conditions, only the LusPAL1 and Lus4CL4 genes were downregulated and 7 genes or gene-groups were significantly upregulated. When taken together, our results showed that the strongest impact on the previously genes identified as involved in lignin biosynthesis, occurred in the stem during dehydration, wounding and in the presence of SA.
Gene clustering
Genes involved in the same biological process are often co-regulated. To further confirm the implication of the former identified genes in the lignification pathway, we searched for possible common expression profiles among the gene set. A k-means clustering approach was used to identify correlated groups exhibiting similar expression profiles in the different tissues and/or stress conditions. The number of flax genes in each cluster ranged between 2 and 14 (Fig. 7a). Among the flax phenylpropanoid associated genes, all the potential bona fide genes identified previously, except for LusCAD1_2 and LusCCR11, were included in cluster 9 (Fig. 7b) showing that the expression of these genes were closely co-regulated in flax. Altogether, the clustering results provide further support for the involvement of the identified genes in lignification.
Fig. 7.
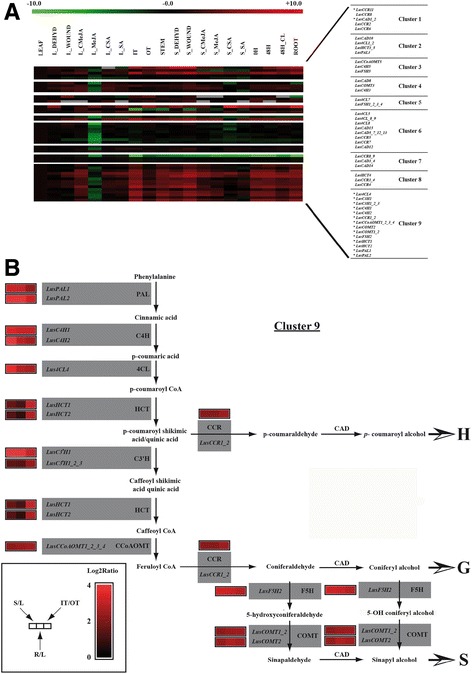
Clustering of flax phenylpropanoid genes according to their expression profile in different organs and stress conditions. a the k-means medians clustering function from the TIGR MultiExperiment Viewer (TM4 Mev v4.8.1) was used to cluster the genes according to their expression profiles. The distance metric was Pearson correlation and the default parameters were used for the k-means calculation. The expression profiles are represented as heat maps. The colour code and expression range in log2 values are represented on top of the figure. b the biosynthetic pathway leading to monolignol production and further to H, G and S units is summarized. Three expression ratios are indicted as heat maps beside the genes. C: control; CL: continuous light; DEHYD: drought stressed stems; IT: internal stem tissues; L: leaf; MeJA: methyljasmonate; OT: outer stem tissues; R: root; S; stem; SA: salicylic acid
Gene co-regulation requires the activity of common transcription factors and since MYB proteins are known to function as master switches of secondary cell wall and lignin gene transcription [50, 51] we examined the promoters of our identified genes for MYB consensus motifs named MBS (MYB binding site) (C/T)AAC(A/T)A(A/C)C and MBSIIG (or SMRE: secondary wall MYB-responsive element) (C/T)ACC(A/T)A(A/C)C. Our results (Additional file 6: Table S3) showed that the MBSIIG site was present in the 500 bp proximal promoter fragments of all the bona fide genes present in the cluster 9 whereas both MBS sites are absent from the LusCCR11 promoter in cluster 1. Interestingly, both MYB sites were absent from the proximal promoters of the LusF5H1_2_3_4 genes which is in accordance with previous results showing that F5H is directly regulated by a NAC SND1 transcription factor [52].
Discussion
Identification of genes involved in the lignin biosynthetic pathway in flax
In the past 25 years, a tremendous amount of information about lignin biosynthesis has been obtained using model plants such as Arabidopsis and Populus species. Currently, the possibility to obtain comprehensive genomic sequences in other species is allowing the scientific community to gather additional data on other species. Secondary cell walls (SCWs), whether they are specialized in water conduction (xylem) or mechanical support (sclerenchyma), usually contain around 25% lignin. However, in some specialized tissues such as bast fibers, the cell wall contains very low amounts of lignin together with high cellulose levels [53, 54]. Although these cells only contain low amounts of lignins, the presence of these phenolic molecules can have an important impact on the quality of the final products such as linen tissues and biocomposites.
In this context, we carried out a comprehensive genome-wide analysis of the gene set involved in phenylpropanoids, and more specifically in lignin biosynthesis in flax. The size of individual phenylpropanoid gene families is difficult to compare between species due to differences in the stringency of the sequence annotations. This is especially true for widespread enzymatic activities such as hydroxylases or methyltransferases. On the other hand, gene expression profiling often gives clear information about the function of the more specific bona fide lignin genes [24]. Usually, individual qRT-PCR experiments are performed to validate lignin transcript level pattern analysis [55] but HT-RT-qPCR approaches are increasingly used as they are more appropriately sized for characterizing the members of multigene families in a single step [56]. In flax, we first identified the genes potentially involved in lignification by examining the expression profiles in roots and stems compared to the leaves and also by comparison between the internal xylem stem tissues (high lignin) and the outer fiber-bearing tissues (low lignin).
Further confirmation of the likely implication of selected phenylpropanoid genes in lignification was provided by in situ hybridization, allowing the precise localization of the lignin biosynthetic gene expression. Both in situ hybridization and also promoter-reporter gene studies conducted on a low number of genes previously showed that bona fide lignin genes were expressed in the first few cell layers of differentiating secondary xylem [17–19, 57, 58]. To our knowledge, our work reports for the first time an in situ hybridization approach on a large number of lignin genes in a higher plant species. As was shown by the results of the high throughput expression analyses, the very high expression levels in the xylem-rich tissues compared to the bast fiber cells is in accordance with our previous results suggesting that lignification of flax fibers is at least in part regulated at the transcriptional level [15].
Finally, when the expression patterns of all the phenylpropanoid genes were considered, most of these lignin-associated genes were found to be co-expressed within a same cluster confirming that these genes were indeed involved in a common metabolic pathway. The presence of common MYB cis-element motifs in the promoters of these genes associated to their co-expression was already described for the orthologous genes in Arabidopsis [59]. In this species, MYB58 can directly activate most lignin biosynthesis genes but not F5H showing that the syringyl lignin biosynthesis is activated by a different regulatory pathway probably controlled by NAC transcription factors [52].
Selection of candidates for lignin engineering in flax
The success of targeted engineering of cell wall lignin genes to improve the quality of different flax-derived products depends upon the correct identification of individual family gene members involved in the trait to be modified. Some genes potentially involved in lignin biosynthesis have already been partially characterized in flax. The first flax lignin cDNA sequence was deposited in the GenBank database in 2005 and the corresponding CCoAOMT gene functionally characterized [30]. It was chosen because it is responsible for the synthesis of feruloyl CoA used for the formation of both G- and S-lignin units and, as confirmed by down-regulation, because it plays a central role in maintaining SCW integrity by regulating the quantity and the S/G proportion in the non-condensed lignin fraction. This gene corresponds to the LusCCoAOMT4 identified in our study and is included in the bona fide group of genes expressed in lignified tissues. In another study, a CAD gene fragment sharing 100% homology with LusCAD4 was targeted by a downregulation strategy [60]. This gene associated with LusCAD3 was not retained in our work as a major lignin gene because of its much lower differential expression between lignified and poorly-lignified organs or tissues when compared to LusCAD1_2. Although the RNAi transformed plants contained lower amounts of lignins compared to the control, it is interesting to note that M Wrobel-Kwiatkowska, M Starzycki, J Zebrowski, J Oszmianski and J Szopa [60]) did not observe the typical brown-midrib phenotype of CAD mutants observed in other species. In contrast, TILLed flax chemical mutants of LusCAD1 possess the characteristic orange-coloured xylem phenotype [16] confirming the likely involvement of this gene in the lignification process. Flax lignin has also received interest because of the very low amounts of S-units [53] so we were therefore interested to examine the F5H gene family. This enzyme is specifically required for the synthesis of S lignin since it catalyses the 5-hydroxylation of coniferaldehyde and/or coniferyl alcohol [35]. Its down-regulation in Arabidopsis leads to a reduced S/G value [61]. The expression ratio of LusF5H2 in the lignified organs and tissues is comparable to those of other identified genes so the biosynthesis of low amounts of S units in flax does not seem to be controlled by F5H gene expression. However, further computational modelling and biochemical characterization of the members of the F5H family have to be done in order to provide more clues on the specificity of these enzymes towards various substrates.
lus-miR397 controls several laccase genes in flax
Laccases are mono- or multimeric copper-oxidases in eukaryotes and procaryotes with a broad range of aromatic or non-aromatic substrates [62] and have been implicated in the oxidative polymerization of monolignols. Among the 17 gene models described in Arabidopsis, AthLAC4, AthLAC11 and AthLAC17 were clearly shown to contribute to the constitutive lignification of the floral stems [43, 63]. In Populus trichocarpa, 49 gene models have been identified but it was not yet possible to clearly identify those involved in lignification [21]. In flax, we identified 45 laccase gene models of which 23 were highly expressed in lignified tissues. MicroRNA targeting (miR397) of some laccase transcripts, including AthLAC4 and AthLAC17, contribute to the overall gene regulation [20, 21]. This microRNA has been identified in many gymnosperms, monocots and dicots with 1 to 3 copies per species [64]. In this study, we experimentally demonstrated that transcripts corresponding to six flax laccase genes were targeted by lus-miR397. On the basis of our phylogenetic analysis, five of these genes were found in the clade 1, which also contained AthLAC4 whereas six non-targeted genes were present in clade 2 along with AthLAC17 showing that either no functional ortholog of this gene is present in flax or that these genes have lost their ability to be cleaved by lus-miR397. Strong evidence for a likely role of microRNA regulation of laccase-mediated lignification in flax was provided by the cell−/tissue-specific localization of the corresponding transcript precursors. Our results are in agreement with those obtained with a GUS-GFP fusion driven by the miR397b promoter, showing that this gene is expressed in the stem vascular tissues of Arabidopsis [65]. Further analyses should allow a better understanding of whether miR397 plays a role in maintaining constant transcript levels or is implicated in a finer control of different laccase paralogs.
Stress related phenylpropanoid genes
Flax plants selected for fiber production are mostly cultivated around the Mediterranean and temperate climate zones. In the context of climate change, cell wall metabolism, structure and fiber quality are likely to be affected in the future and it is therefore informative to know which genes are affected by biotic or abiotic stresses. We took advantage of the high throughput gene expression approach to try and distinguish between developmental lignin genes and stress-related phenylpropanoid gene expression.
Defence-related hormones such as SA and MeJA can control lignin gene activation. SA is an important signalling molecule for systemic acquired resistance [66] and triggers the expression of lignin genes in many different species [67]. In flax, several lignin genes showed opposite expression profiles in the presence of SA suggesting that the composition of lignin may change during the defence mechanism or that more specific metabolic pathways including these genes are activated. The effect of MeJA on this metabolic pathway seems more subtle. This component has been shown to participate in the signal transduction pathway in response to different forms of stresses [68]. A dose effect was described in Fragaria fruits [69] as well as an opposite CAD gene expression pattern in tea plants [70]. In our experimental conditions, most of the flax genes were underexpressed indicating that the synthesis of lignin is probably not part of the response to this elicitor. Abiotic stresses can also have an influence on lignification. Mechanical stresses can be responsible for the deposition of various polymers including lignin that may function as a physical barrier around the injury zone [71]. In this context, lignin genes are activated in different species including sweet potato [72], maize [73] or Arabidopsis [74]. Most of the flax lignin genes were activated in the stem but less in the leaves showing that in this species, lignin is also synthesized in reaction to wounding. Outsides these bona fide genes, several common phenylpropanoid genes including CADs were also activated in both organs showing that they may potentially be involved in the production of other abiotic stress responsive metabolites as was already proposed in poplar [38]. The effect of drought stress on plants is more difficult to understand because it can have positive or negative roles on lignification depending on the species, the organs/tissues and the intensity and duration of the stress period [75]. In our experimental conditions (12 days after watering stop) most of the lignin genes were upregulated in the stems and the leaves. The increase in lignification has been described as one of the reactions included in a general adaptation strategy of plants faced with water loss and may result in an increase of mechanical strength and/or water impermeability [76].
Conclusion
We have identified, in this study, the individual members of the multigenic families most likely involved in lignin biosynthesis (and polymerisation) in flax. These data provide the knowledge base necessary to undertake targeted engineering and/or marker-based selection of lignin biosynthesis in flax, as well as important information on the behaviour of these genes in a number of different stress conditions. The latter information will be particularly useful for farmers and breeders in the current context of increasing variability in weather conditions associated with climate change.
Methods
Plant material
For HT-RT-qPCR expression analysis, flax plants (Linum usitatissimum L. cv Diane) were grown in 12 × 12 cm soil-containing pots in a greenhouse under 16 h/20 °C day and 8 h/18 °C night conditions for 35 days. For developmental expression studies, (i) leaves, ii) 2-cm root fragments taken under the stem base, (iii) top 4 cm of stem, (iv) fiber-rich outer-tissues and (v) xylem-rich inner-tissues from stem base were harvested. For stress expression studies, the following samples were taken from these 35-day-old plants: (i) leaves and stems harvested 24 h after wounding with a scalpel, (ii) leaves and stems incubated for 24 h in Petri dishes containing 200 μM salicylic acid (SA) or methyljasmonate (MeJA) dissolved in ½ MS medium [77], (iii) leaves and stems from plants submitted to a drought stress imposed by withholding water for 12 days, (iv) stems from plants first placed in phytotrons during 7 days for acclimation and then submitted to continuous light for 48 h at 20 °C. All the samples were triplicates and always frozen immediately in liquid nitrogen before storage at −80 °C prior to RNA extraction. For in situ hybridization, the median stem, leaves and 2-cm root fragments were collected and fixed as described below.
In silico identification and analysis of phenylpropanoid genes in L. usitatissimum
The flax phenylpropanoid protein and gene sequences were identified in the annotated v1.0 of the Linum usitatissimum genome hosted on the Phytozome website [45]. The entire database derived from the published genome sequences [10] was queried with the names of the gene families used as keywords and also by BLASTp interrogation with previously lignin-related identified sequences from Arabidopsis thaliana [78] and Populus trichocarpa [25]. When it was necessary, unknown portions of some sequences were replaced by the corresponding sequences found in public EST databases and truncated sequences were discarded. For each gene model, the presence of a corresponding EST was searched in the dbEST [79] and genolin [11] databases. The intron/exon structures were also retrieved and the identities of the protein sequences compared. The k-means medians clustering function from the TIGR MultiExperiment Viewer (TM4 Mev v4.8.1) was used to cluster the genes according to their expression profiles. The distance metric was Pearson correlation and the default parameters were used for the k-means calculation.
Phylogenetic analyses
For each gene family, the protein sequences were aligned with ClustalW and the phylogenetic analyses obtained with the MEGA 6.06 [80] software for performed by maximum likelihood method based on the JTT matrix-based model [81]. Consensus trees were generated with 1000 bootstrap replicates.
HT-RT-qPCR using a 96.96 dynamic array
Total RNA was extracted from the different organs and tissues using the TriReagent method (Molecular Research Center). RNA integrity and concentration were evaluated with RNA Standard Sens Chips in the Experion automated capillary electrophoresis system (Bio-Rad). cDNA was then synthesized using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s instructions. The large scale quantitative RT-PCR was performed with a BioMark HD System using a Fluidigm 96.96 dynamic array (IntegraGen, Evry France) according to the Fluidigm Advanced Development Protocol with EvaGreen using primer pairs listed in Additional file 4: Table S1. A first preamplification reaction (1 cycle: 95 °C 10 min; 14 cycles: 95 °C 15 s, 60 °C 4 min) was performed for each sample in 10 μl by pooling primer pairs (final concentration, 50 nM), 3.3 μl cDNA, and 5 μl 2X PreAmp Master Mix (Applied Biosystems). For each assay, 5 μl 10X Assay Mix containing 9 μM forward primer, 9 μM reverse primer, and 1X Assay Loading Reagent was loaded into the chip. The following thermal cycles were executed: 1 cycle: 95 °C 10 min; 40 cycles: 95 °C 15 s, 60 °C 1 min. The amplifications on the three biological samples were always conducted in triplicate and were also performed on a mix of cDNAs to determine the amplification efficiencies. The Ct values were analysed with the qBase + software (Biogazelle, Belgium). The data normalization was performed using the previous experimentally validated reference genes LusETIF5A1, LusUBI1 and LusEF1A [82].
Histochemical analysis
Flax stems, roots and leaves were fixed with 4% paraformaldehyde in phosphate buffer (0.2 M, pH 7) for 16 h. They were then washed in phosphate buffer containing 0.1 M glycine and dehydrated through a series of ethanol solutions and progressively embedded in paraffin (ParaplastPlus; Oxford Labware). Tissue sections were obtained with a Leica RM2065 microtome and placed on silanised-coated slides. Before staining, tissue sections were deparaffinised with Histoclear (Labonord) and rehydrated with decreasing concentrations of ethanol. The presence of lignin was determined by staining with the Wiesner reagent (phloroglucinol–HCl) [83].
Spatial gene expression determination by in situ hybridization
To obtain DNA templates for RNA probe synthesis, PCR amplifications were performed with gene-specific primers tailed with a T7 RNA polymerase binding site Additional file 4: Table S1. The amplicons (1 μg) were used as templates to synthesize sense and antisense probes for each gene, with the incorporation of UTP–digoxigenin as the label using the DIG labelling mix (Roche). Deparaffinised sections were treated as previously described [84], hydridized and probe detected with NBT (nitro-blue tetrazolium chloride)/BCIP (5-bromo-4-3′-indolylphosphate p-toluidine salts) solutions according to the manufacturer’s instructions. Controls without probe or with sense probe were performed to check for endogenous alkaline phosphate activity.
miRNA target prediction and experimental validation
The sequence of the lus-miR397 precursor was identified among the known flax ESTs using stringent rules described elsewhere [44]. The laccase transcript models were analysed with psRNATarget [85] to predict the targets that can potentially match with the microRNA without any gaps. When referring to the miRNA sequence, there were not more than one mismatch between the nucleotides number 1 and 9, none between 10 and 11, and no more than 2 consecutive mismatches after position 11. The experimental validation of the laccase targets was carried out by using a modified protocol of the RNA ligation mediated (RLM) RACE included in the GeneRacer kit (Invitrogen). RNA was extracted from a mix of flax organs and the cleaved transcripts (without 5′-cap) were selectively ligated to a 5′-RNA adaptor. After reverse transcription, PCR amplification was performed with a 5′ adaptor-specific primer and a reverse gene specific primer Additional file 4: Table S1 located downstream of the predicted cleavage site. The fragments were then gel purified and sequenced.
Additional files
Flax phenylpropanoid gene structures. The name of the gene is indicated followed by the length in bp between the start and stop codons in parentheses. The exons are shown as boxes and the introns as lines. The scale in bp is shown on the top of the figure. (DOC 40 kb)
Molecular phylogenetic analysis of the phenylpropanoid genes. (PDF 1379 kb)
Expression of the flax phenylpropanoid genes under stress conditions as determined by HT-RT-qPCR. 0H: start of the light period on the first day; 48H: end of the night on the second day; 48H_CL: 48 h after continuous light; CONT: control; Dehyd: dehydration; L: leaf; MeJA: methyljasmonate; S: stem; SA: salicylic acid. (PDF 1906 kb)
Primers used for HT-RT-qPCR, RLM RACE and in situ hybridization. (DOC 76 kb)
Description of the gene expression localization in the tissues of flax roots, stems and leaves. (JPEG 391 kb)
Position of the MBSII and MBSIIG specific cis elements on both strands (+/−) of the flax phenylpropanoid gene promoters. (XLSX 15 kb)
Acknowledgements
We are indebted to the Research Federation FRABio (Univ. Lille, CNRS, FR 3688, FRABio, Biochimie Structurale et Fonctionnelle des Assemblages Biomoléculaires) for providing the scientific and technical environment conducive to achieving this work. JLR gratefully acknowledges the french Ministère de l’Enseignement Supérieur et de la Recherche for financial support.
Funding
This research was supported by the french Ministère de l’Enseignement Supérieur et de la Recherche.
Availability of data and materials
The flax DNA sequences mentioned in this article can all be retrieved from the Phytozome database: https://phytozome.jgi.doe.gov/pz/portal.html.
Abbreviations
- 4CL
4-coumarate:CoA ligase
- C3’H
p-coumarate 3-hydroxylase
- C4H
cinammate 4-hydroxylase
- CAD
cinnamyl alcohol dehydrogenase
- CCoAOMT
caffeoyl CoA 3-O-methyltransferase
- CCR
cinnamoyl CoA reductase
- COMT
caffeate/5-hydroxyferulate O-methyl-transferase
- F5H
ferulate 5-hydroxylase
- HCT
hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase
- MeJA
methyljasmonate
- PAL
Phenylalanine ammonia-lyase
- SA
salicylic acid
Authors’ contributions
JLR and ASB performed the sample preparations and experimental procedures, GN participated in the experiments, AC performed data analysis, BH and SH critically revised the manuscript and GN designed the study, supervised the work and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable..
Consent for publication
Not applicable..
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-1072-9) contains supplementary material, which is available to authorized users.
References
- 1.Le Roy J, Huss B, Creach A, Hawkins S, Neutelings G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci. 2016;7 [DOI] [PMC free article] [PubMed]
- 2.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 3.Freudenberg K. Lignin biosynthesis. Berlin, Heidelberg, New York: Springer; 1968. [Google Scholar]
- 4.Sterjiades R, Dean JF, Eriksson KE. Laccase from sycamore maple (Acer pseudoplatanus) polymerizes Monolignols. Plant Physiol. 1992;99(3):1162–1168. doi: 10.1104/pp.99.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moura JCMS, Bonine CAV. De Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol. 2010;52(4):360–376. doi: 10.1111/j.1744-7909.2010.00892.x. [DOI] [PubMed] [Google Scholar]
- 6.Neutelings G. Lignin variability in plant cell walls: contribution of new models. Plant Sci. 2011;181(4):379–386. doi: 10.1016/j.plantsci.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Mohanty AK, Misra M, Hinrichsen G. Biofibres, biodegradable polymers and biocomposites: an overview. Macromol Mater Eng. 2000;276-277(1):1–24. doi: 10.1002/(SICI)1439-2054(20000301)276:1<1::AID-MAME1>3.0.CO;2-W. [DOI] [Google Scholar]
- 8.Zohary D, Hopf M. Domestication of plants in the old world, third edition edn. New York: Oxford University Press; 2004. [Google Scholar]
- 9.Summerscales J, Dissanayake NPJ, Virk AS, Hall W. A review of bast fibres and their composites. Part 1 – Fibres as reinforcements. Compos A: Appl Sci Manuf. 2010;41(10):1329–1335. doi: 10.1016/j.compositesa.2010.06.001. [DOI] [Google Scholar]
- 10.Wang Z, Hobson N, Galindo L, Zhu S, Shi D, McDill J, Yang L, Hawkins S, Neutelings G, Datla R, et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012;72(3):461–473. doi: 10.1111/j.1365-313X.2012.05093.x. [DOI] [PubMed] [Google Scholar]
- 11.Fenart S, Ndong Y-P, Duarte J, Riviere N, Wilmer J, van Wuytswinkel O, Lucau A, Cariou E, Neutelings G, Gutierrez L. Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics. 2010;11(1):592. doi: 10.1186/1471-2164-11-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chantreau M, Chabbert B, Billiard S, Hawkins S, Neutelings G. Functional analyses of cellulose synthase genes in flax (Linum usitatissimum) by virus-induced gene silencing. Plant Biotechnol J. 2015;13(9):1312–1324. doi: 10.1111/pbi.12350. [DOI] [PubMed] [Google Scholar]
- 13.Ragupathy R, Rathinavelu R, Cloutier S. Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome. BMC Genomics. 2011;12:217. doi: 10.1186/1471-2164-12-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day A, Fenart S, Neutelings G, Hawkins S, Rolando C, Tokarski C. Identification of cell wall proteins in the flax (Linum usitatissimum) stem. Proteomics. 2013;13(5):812–825. doi: 10.1002/pmic.201200257. [DOI] [PubMed] [Google Scholar]
- 15.Huis R, Morreel K, Fliniaux O, Lucau-Danila A, Fenart S, Grec S, Neutelings G, Chabbert B, Mesnard F, Boerjan WA. Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol. 2012;158(4):1893–1915. doi: 10.1104/pp.111.192328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chantreau M, Grec S, Gutierrez L, Dalmais M, Pineau C, Demailly H, Paysant-Leroux C, Tavernier R, Trouve JP, Chatterjee M, et al. PT-flax (phenotyping and TILLinG of flax): development of a flax (Linum usitatissimum L.) mutant population and TILLinG platform for forward and reverse genetics. BMC Plant Biol. 2013;13:159. doi: 10.1186/1471-2229-13-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding SA, Leshkevich J, Chiang VL, Tsai CJ. Differential substrate inhibition couples kinetically distinct 4-Coumarate:coenzyme a ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol. 2002;128(2):428–438. doi: 10.1104/pp.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins S, Boudet A, Grima Pettenati J. Characterisation of caffeic acid O-methyltransferase and cinnamyl alcohol dehydrogenase gene expression patterns by in situ hybridisation in Eucalyptus gunnii hook. Plantlets. Plant Sci. 2003;164(2):165–173. doi: 10.1016/S0168-9452(02)00361-8. [DOI] [Google Scholar]
- 19.Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, Boudet AM, Grima-Pettenati J. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 1997;11(3):429–441. doi: 10.1046/j.1365-313X.1997.11030429.x. [DOI] [PubMed] [Google Scholar]
- 20.Abdel Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem. 2008;283(23):15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu S, Li Q, Wei H, Chang MJ, Tunlaya Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proceedings of the Natlional Academy of Science USA. 2013;110(26):10848–10853. doi: 10.1073/pnas.1308936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chantreau M, Portelette A, Dauwe R, Kiyoto S, Cronier D, Morreel K, Arribat S, Neutelings G, Chabi M, Boerjan W, et al. Ectopic lignification in the flax lignified bast fiber1 mutant stem is associated with tissue-specific modifications in gene expression and Cell Wall composition. Plant Cell. 2014;26(11):4462–4482. doi: 10.1105/tpc.114.130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roach MJ, Deyholos MK. Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol Gen Genomics. 2007;278(2):149–165. doi: 10.1007/s00438-007-0241-1. [DOI] [PubMed] [Google Scholar]
- 24.Carocha V, Soler M, Hefer C, Cassan-Wang H, Fevereiro P, Myburg AA, Paiva JA, Grima-Pettenati J. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytologist. 2015;206(4):1297–1313. doi: 10.1111/nph.13313. [DOI] [PubMed] [Google Scholar]
- 25.Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010;51(1):144–163. doi: 10.1093/pcp/pcp175. [DOI] [PubMed] [Google Scholar]
- 26.Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell. 2004;16(10):2749–2771. doi: 10.1105/tpc.104.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Azevedo SC, Barbazuk B, Ralph SG, Bohlmann J, Hamberger B, Douglas CJ. Genome-wide analysis of a land plant-specific acyl:coenzymeA synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol. 2008;179(4):987–1003. doi: 10.1111/j.1469-8137.2008.02534.x. [DOI] [PubMed] [Google Scholar]
- 28.Ehlting J, Buttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme a ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19(1):9–20. doi: 10.1046/j.1365-313X.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 29.Hamberger B, Ellis M, Friedmann M, de Azevedo SC, Barbazuk B, Douglas CJ. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot. 2007;85(12):1182–1201. doi: 10.1139/B07-098. [DOI] [Google Scholar]
- 30.Day A, Neutelings G, Nolin F, Grec S, Habrant A, Cronier D, Maher B, Rolando C, David H, Chabbert B, et al. Caffeoyl coenzyme a O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem. 2009;47(1):9–19. doi: 10.1016/j.plaphy.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Barakat A, Yassin NB, Park JS, Choi A, Herr J, Carlson JE. Comparative and phylogenomic analyses of cinnamoyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants. Plant Sci. 2011;181(3):249–257. doi: 10.1016/j.plantsci.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Bugos RC, Chiang VL, Campbell WH. cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol Biol. 1991;17(6):1203–1215. doi: 10.1007/BF00028736. [DOI] [PubMed] [Google Scholar]
- 33.Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proceedings of the Natlional Academy of Science USA. 1999;96(16):8955–8960. doi: 10.1073/pnas.96.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P et al. Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 2009; 10 Suppl 11:S3. [DOI] [PMC free article] [PubMed]
- 35.Humphreys JM, Hemm MR, Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proceedings of the Natlional Academy of Science USA. 1999;96(18):10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansell RE, Stöckigt J, Zenk H. Reduction of Ferulic acid to Coniferyl alcohol in a cell free system from a higher plant. Z Pflanzenphysiol. 1972;68:286–288. doi: 10.1016/S0044-328X(72)80063-1. [DOI] [Google Scholar]
- 37.Guo DM, Ran JH, Wang XQ. Evolution of the Cinnamyl/Sinapyl alcohol dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of Bona fide CAD. J Mol Evol. 2010;71(3):202–218. doi: 10.1007/s00239-010-9378-3. [DOI] [PubMed] [Google Scholar]
- 38.Bagniewska Zadworna A, Barakat A, Lakomy P, Smolinski DJ, Zadworny M. Lignin and lignans in plant defence: insight from expression profiling of cinnamyl alcohol dehydrogenase genes during development and following fungal infection in Populus. Plant Sci. 2014;229:111–121. doi: 10.1016/j.plantsci.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Chao N, Liu SX, Liu BM, Li N, Jiang XN, Gai Y. Molecular cloning and functional analysis of nine cinnamyl alcohol dehydrogenase family members in Populus tomentosa. Planta. 2014;240(5):1097–1112. doi: 10.1007/s00425-014-2128-9. [DOI] [PubMed] [Google Scholar]
- 40.Jin Y, Zhang C, Liu W, Qi H, Chen H, Cao S. The cinnamyl alcohol dehydrogenase gene family in melon (Cucumis melo L.): bioinformatic analysis and expression patterns. PLoS One. 2014;9(7):e101730. doi: 10.1371/journal.pone.0101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics. 2009;181(2):783–795. doi: 10.1534/genetics.108.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Parijs FR, Ruttink T, Boerjan W, Haesaert G, Byrne SL, Asp T, Roldan-Ruiz I, Muylle H. Clade classification of monolignol biosynthesis gene family members reveals target genes to decrease lignin in Lolium perenne. Plant Biology (Stuttg) 2015;17(4):877–892. doi: 10.1111/plb.12316. [DOI] [PubMed] [Google Scholar]
- 43.Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cezard L, Le Bris P, Borrega N, Herve J, Blondet E, Balzergue S, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23(3):1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neutelings G, Fenart S, Lucau-Danila A, Hawkins S. Identification and characterization of miRNAs and their potential targets in flax. J Plant Physiol. 2012;169(17):1754–1766. doi: 10.1016/j.jplph.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu S, Yang C, Chiang VL. Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. J Integr Plant Biol. 2011;53(11):879–891. doi: 10.1111/j.1744-7909.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 47.Lu H. Dissection of salicylic acid-mediated defense signaling networks. Plant Signal Behav. 2009;4(8):713–717. doi: 10.4161/psb.4.8.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M, Mansfield J, Zipfel C, Hamann T. Cell Wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and Jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 2011;156(3):1364–1374. doi: 10.1104/pp.111.175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim EH, Kim YS, Park SH, Koo YJ, Choi YD, Chung YY, Lee IJ, Kim JK. Methyl Jasmonate reduces grain yield by mediating stress signals to Alter spikelet development in Rice. Plant Physiol. 2009;149(4):1751–1760. doi: 10.1104/pp.108.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahantamalala A, Rech P, Martinez Y, Chaubet-Gigot N, Grima-Pettenati J, Pacquit V. Coordinated transcriptional regulation of two key genes in the lignin branch pathway--CAD and CCR--is mediated through MYB- binding sites. BMC Plant Biol. 2010;10:130. doi: 10.1186/1471-2229-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong R, McCarthy RL, Haghighat M, Ye ZH. The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0069219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Q, Wang H, Yin Y, Xu Y, Chen F, Dixon RA. Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proceedings of the Natlional Academy of Science USA. 2010;107(32):14496–14501. doi: 10.1073/pnas.1009170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Day A, Ruel K, Neutelings G, Crônier D, David H, Hawkins S, Chabbert B. Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 2005; 222(2):234–45. [DOI] [PubMed]
- 54.Lapierre C, Pollet B, Petit Conil M, Toval G, Romero J, Pilate G, Leplé JC, Boerjan W, Ferret V, De Nadai V, et al. Structural alterations of Lignins in transgenic poplars with depressed Cinnamyl alcohol dehydrogenase or Caffeic acid O-Methyltransferase activity have an opposite impact on the efficiency of industrial Kraft pulping. Plant Physiol. 1999;119(1):153–164. doi: 10.1104/pp.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen H, Mazarei M, Hisano H, Escamilla-Trevino L, Fu C, Pu Y, Rudis MR, Tang Y, Xiao X, Jackson L, et al. A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell. 2013;25(11):4342–4361. doi: 10.1105/tpc.113.118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinzon-Latorre D, Deyholos MK. Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum) BMC Genomics. 2013;14:742. doi: 10.1186/1471-2164-14-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feuillet C, Lauvergeat V, Deswarte C, Pilate G, Boudet A, Grima-Pettenati J. Tissue- and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol Biol. 1995;27(4):651–667. doi: 10.1007/BF00020220. [DOI] [PubMed] [Google Scholar]
- 58.Hawkins S, Samaj J, Lauvergeat V, Boudet A, Grima-Pettenati J. Cinnamyl alcohol dehydrogenase: identification of new sites of promoter activity in transgenic poplar. Plant Physiol. 1997;113(2):321–325. doi: 10.1104/pp.113.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma S, Shah S, Bohnert HJ, Snyder M, Dinesh Kumar SP. Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet. 2013;9(10) doi: 10.1371/journal.pgen.1003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wrobel-Kwiatkowska M, Starzycki M, Zebrowski J, Oszmianski J, Szopa J. Lignin deficiency in transgenic flax resulted in plants with improved mechanical properties. J Biotechnol. 2007;128(4):919–934. doi: 10.1016/j.jbiotec.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 61.Ohman D, Demedts B, Kumar M, Gerber L, Gorzsas A, Goeminne G, Hedenstrom M, Ellis B, Boerjan W, Sundberg B. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 2013;73(1):63–76. doi: 10.1111/tpj.12018. [DOI] [PubMed] [Google Scholar]
- 62.Berthet S, Thevenin J, Baratiny D, Demont-Caulet N, Debeaujon I, Przemyslaw B, Leplé J, Huis R, Hawkins S, Gomez LD, et al. Role of plant Laccases in lignin polymerization. Chapter 5. Adv Bot Res. 2012;61:145–172. doi: 10.1016/B978-0-12-416023-1.00005-7. [DOI] [Google Scholar]
- 63.Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25(10):3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang CY, Zhang S, Yu Y, Luo YC, Liu Q, Ju C, Zhang YC, Qu LH, Lucas WJ, Wang X, et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol J. 2014;12(8):1132–1142. doi: 10.1111/pbi.12222. [DOI] [PubMed] [Google Scholar]
- 66.Yalpani N, Raskin I. Salicylic acid: a systemic signal in induced plant disease resistance. Trends Microbiology. 1993;1(3):88–92. doi: 10.1016/0966-842X(93)90113-6. [DOI] [PubMed] [Google Scholar]
- 67.Miedes E, Boerjan W, Vanholme R, Molina A. The role of the secondary cell walls in plant resistance to pathogens. Front Plant Sci. 2014;5 [DOI] [PMC free article] [PubMed]
- 68.Sharma M, Laxmi A. Jasmonates: emerging players in controlling temperature stress tolerance. Front Plant Sci. 2015;6:1129. doi: 10.3389/fpls.2015.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Concha CM, Figueroa NE, Poblete LA, Onate FA, Schwab W, Figueroa CR. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol Biochem. 2013;70:433–444. doi: 10.1016/j.plaphy.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Deng WW, Zhang M, Wu JQ, Jiang ZZ, Tang L, Li YY, Wei CL, Jiang CJ, Wan XC. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant. Camellia sinensis Journal of Plant Physiology. 2013;170(3):272–82. [DOI] [PubMed]
- 71.Hawkins S, Boudet A. Wound-induced lignin and suberin deposition in a woody angiosperm (Eucalyptus gunnii hook.): Histochemistry of early changes in young plants. Protoplasma. 1996;191(1):96–104. doi: 10.1007/BF01280829. [DOI] [Google Scholar]
- 72.Kim YH, Bae JM, Huh GH. Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress. Plant Cell Rep. 2010;29(7):779–791. doi: 10.1007/s00299-010-0864-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Obeso M, Caparros-Ruiz D, Vignols F, Puigdomenech P, Rigau J. Characterisation of maize peroxidases having differential patterns of mRNA accumulation in relation to lignifying tissues. Gene. 2003;309(1):23–33. doi: 10.1016/S0378-1119(03)00462-1. [DOI] [PubMed] [Google Scholar]
- 74.Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R. Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol. 2004;55(2):165–181. doi: 10.1007/s11103-004-0112-7. [DOI] [PubMed] [Google Scholar]
- 75.Cabané M, Afif D, Hawkins S. Advances in Botanical Research. In: Lignins and Abiotic Stresses, vol. 61: London: Academic Press Elsevier; 2012. p. 220–62.
- 76.Hu Y, Li WC, Xu YQ, Li GJ, Liao Y, Fu FL. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J Appl Genet. 2009;50(3):213–223. doi: 10.1007/BF03195675. [DOI] [PubMed] [Google Scholar]
- 77.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15.
- 78.Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133(3):1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boguski M, Lowe T, Tolstoshev C. dbEST database for expressed sequence tags. Nat Genet. 1993;4(4):332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 80.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 82.Huis R, Hawkins S, Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.) BMC Plant Biol. 2010;10:71. doi: 10.1186/1471-2229-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clifford MN. Specificity of acidic phloroglucinol reagents. J Chromatogr A. 1974;94:321–324. doi: 10.1016/S0021-9673(01)92389-1. [DOI] [Google Scholar]
- 84.Roongsattham P, Morcillo F, Jantasuriyarat C, Pizot M, Moussu S, Jayaweera D, Collin M, Gonzalez-Carranza ZH, Amblard P, Tregear JW, et al. Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol. 2012;12:150. doi: 10.1186/1471-2229-12-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research 2011; 39(Web Server issue):W155-W159. [DOI] [PMC free article] [PubMed]
- 86.Babu PR, Rao KV, Reddy VD. Structural organization and classification of cytochrome P450 genes in flax (Linum usitatissimum L.) Gene. 2013;513(1):156–162. doi: 10.1016/j.gene.2012.10.040. [DOI] [PubMed] [Google Scholar]
- 87.Day A, Addi M, Kim W, David H, Bert F, Mesnage P, Rolando C, Chabbert B, Neutelings G, Hawkins S. ESTs from the fibre bearing stem tissues of flax (Linum usitatissimum L.): expression analyses of sequences related to Cell Wall development. Plant Biol. 2005;7(1):23–32. doi: 10.1055/s-2004-830462. [DOI] [PubMed] [Google Scholar]
- 88.Venglat P, Xiang D, Qiu S, Stone SL, Tibiche C, Cram D, Alting-Mees M, Nowak J, Cloutier S, Deyholos M, et al. Gene expression analysis of flax seed development. BMC Plant Biol. 2011;11:74. doi: 10.1186/1471-2229-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flax phenylpropanoid gene structures. The name of the gene is indicated followed by the length in bp between the start and stop codons in parentheses. The exons are shown as boxes and the introns as lines. The scale in bp is shown on the top of the figure. (DOC 40 kb)
Molecular phylogenetic analysis of the phenylpropanoid genes. (PDF 1379 kb)
Expression of the flax phenylpropanoid genes under stress conditions as determined by HT-RT-qPCR. 0H: start of the light period on the first day; 48H: end of the night on the second day; 48H_CL: 48 h after continuous light; CONT: control; Dehyd: dehydration; L: leaf; MeJA: methyljasmonate; S: stem; SA: salicylic acid. (PDF 1906 kb)
Primers used for HT-RT-qPCR, RLM RACE and in situ hybridization. (DOC 76 kb)
Description of the gene expression localization in the tissues of flax roots, stems and leaves. (JPEG 391 kb)
Position of the MBSII and MBSIIG specific cis elements on both strands (+/−) of the flax phenylpropanoid gene promoters. (XLSX 15 kb)
Data Availability Statement
The flax DNA sequences mentioned in this article can all be retrieved from the Phytozome database: https://phytozome.jgi.doe.gov/pz/portal.html.


