Fig. 3.
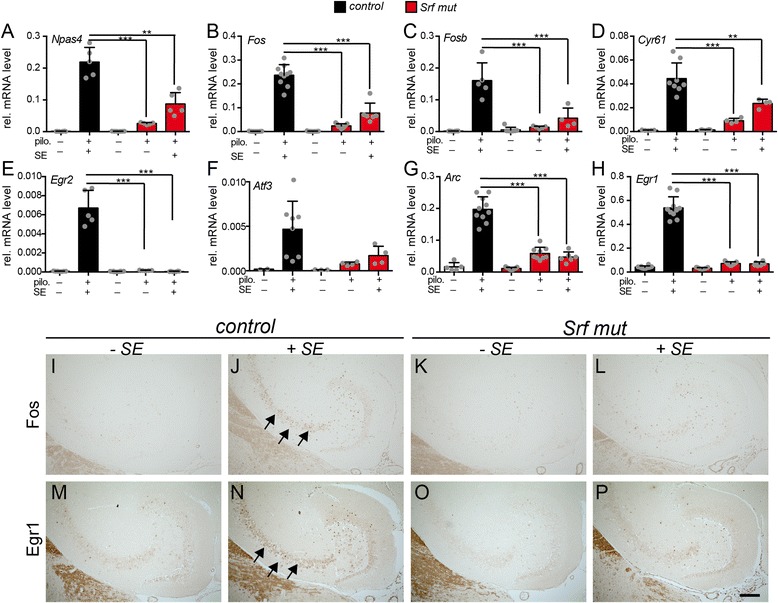
Validation of SRF mediated gene regulation during SE. a-h qPCR validation of SE microarray results with cDNAs derived from independent hippocampal samples. Pilocarpine injected (+pilo) Srf mutant animals either reached SE (+SE) or not (−SE). In heterozygous mice with SE, all IEGs, Fos (b), Fosb (c), Cyr61 (d), Egr2 (e), Arc (g) and Egr1 (h) were upregulated during a 40 min SE, whereas this was significantly reduced in SRF deficient mice with SE. The same holds true for two other genes, Npas4 (a) and Atf3 (f). Data are represented as mean ± SD. Individual animals are labeled with grey circles. i-p Hippocampal sections were labeled with Fos (i-l) or Egr1 (m-p) directed antibodies. In the absence of an SE, neither Fos (i, k) nor Egr1 (m, o) were present in heterozygous (i, m) or Srf mutant (k, o) hippocampal tissues. In control animals with an SE, Fos (j) and Egr1 (n) were upregulated in the CA3 region (see arrows in j, n) and dentate gyrus (DG) on the protein level, whereas this was less pronounced in SRF deficient animals (l, p). Scale-bar (i-p) = 100 μm
