Fig. 6.
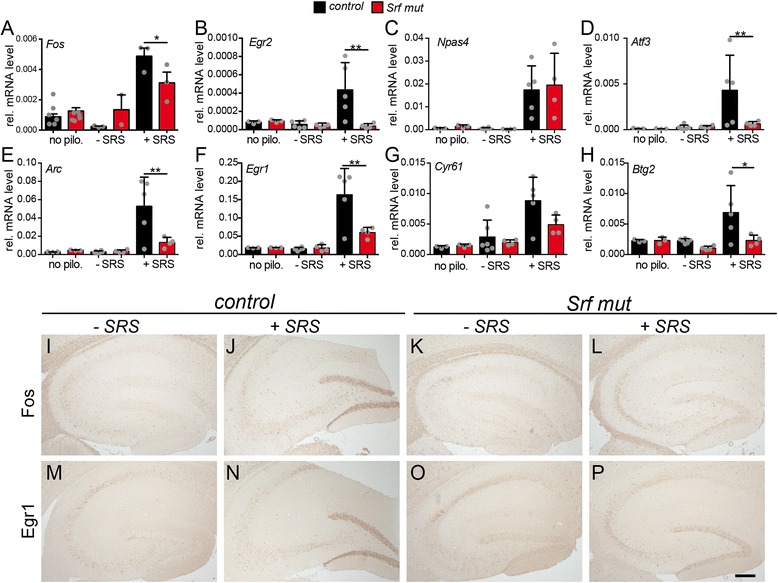
Validation of SRF mediated gene regulation during SRS. a-h qPCR validation of SRS microarray results with cDNAs derived from independent hippocampal samples. Experiments included animals with an SRS (+SRS) and, as control, we included mice with no pilocarpine injection (no pilo.) and epileptic mice not experiencing an SRS within the last 3 h (−SRS). The last two samples showed almost identical mRNA levels for all genes tested. In contrast, 1 h after an SRS, Fos (a), Egr2 (b), Atf3 (d), Arc (e), Egr1 (f), Cyr61 (g) and Btg2 (h) were induced in heterozygous mice. This induction required SRF activity as shown by reduced mRNA levels in Srf mutant animals. In contrast, Npas4 (c) was induced by an SRS, however not in an SRF dependent manner. Data are represented as mean ± SD. Individual animals are labeled with grey circles. For clarity, statistical significance is only depicted for conditions –SRS and +SRS of heterozygous and Srf mut mice. i-p Hippocampal sections of control mice and mice with an SRS were labeled with Fos (i-l) or Egr1 (m-p) directed antibodies. In the absence of an SRS, only weak Fos (i, k) or Egr1 (m, o) expression was present in heterozygous (i, m) or Srf mutant (k, o) hippocampi. In heterozygous animals with an SRS, Fos (j) and Egr1 (n) were upregulated in the hippocampal dentate gyrus, whereas this was decreased in SRF deficient animals (l, p). Scale-bar (i-p) = 100 μm
