Fig. 2.
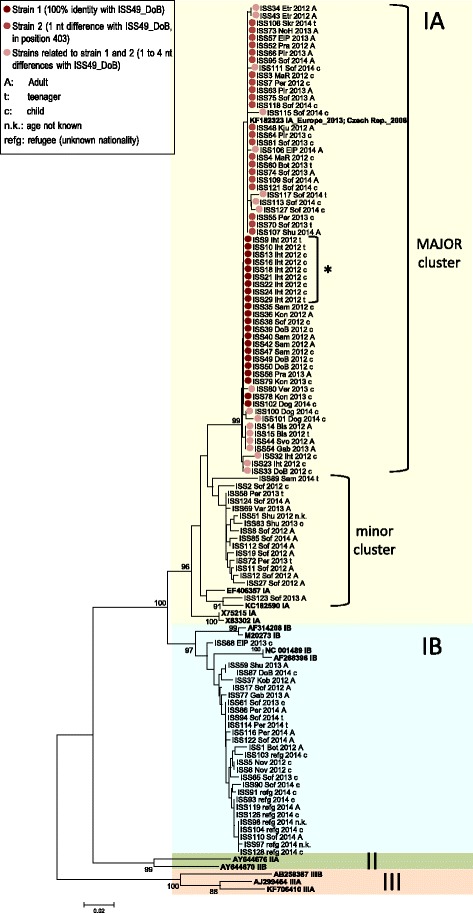
Neighbour-Joining phylogenetic tree (Substitution model: T92 + G) of the 105 HAV isolates from Bulgaria. For each sequence, the town/village (Bis: Bistritsa; Bot: Botevgrad; DoB: Dolna Banja; Dog: Doganovo; ElP: Elin Pelin; Etr: Etropole; Gab: Gabrovo; Iht: Ihtiman; Kju: Kjustendil; Kob: Kostinbrod; Kon: Kostenets; MaR: Manaselska Reka; NoH: Novi Han; Nov: Novachene; Per: Pernik; Pir: Pirdop; Pra: Pravetz; Sam: Samokov; Shu: Shumen; Skr: Skravena; Sof: Sofia; Svo: Svoge; Var: Varna; Ver: Verinsko; refg: refugee, village information not available), the isolation year (2012, 2013, 2014) and the age group of the patient (A: Adult; t: teenager; c: child; n.k.: not known) are reported after the sequence ID. Reference strains with known genotype (IA, IB, IIA, IIB, IIIA, IIIB) were included in the analysis and are reported (in bold) by Accession number followed by the sub-genotype they belong to. Significant bootstrap values are reported. The asterisk shows a small outbreak of 9 cases from Ihtiman occurred between 25 June and 8 August 2012 and involving 6 children and 3 teenagers: the complete nucleotide identity of the isolates confirms a local small outbreak caused by the same strain
