Fig. 5.
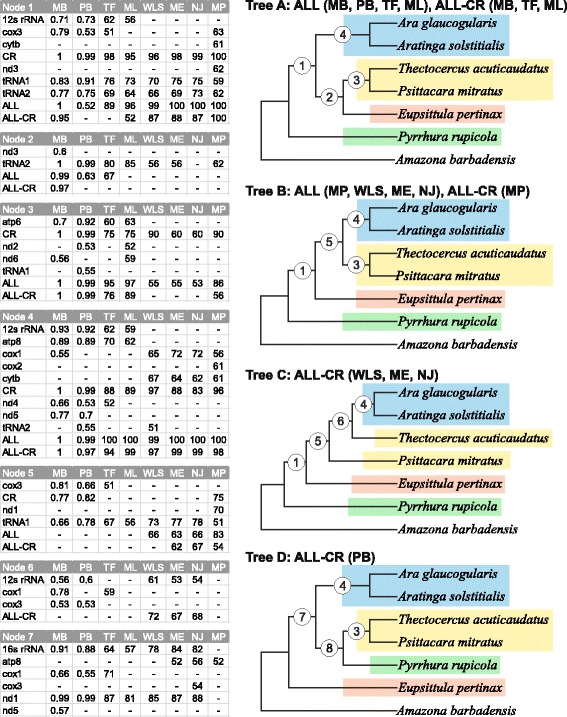
Tree topologies with 7 taxa (A, B, C and D) obtained from concatenated alignments including all markers (ALL) and without the control region (ALL-CR) for the following phylogenetic approaches: Bayesian in MrBayes (MB) and PhyloBayes (PB), maximum likelihood with partitioned data in TreeFinder (TF) and not-partitioned in PAUP (ML) as well as neighbour joining (NJ), minimum evolution (ME), weighted least squares (WLS) and maximum parsimony (MP) in PAUP. The abbreviations of the data sets and methods that produced the given topology are shown above the corresponding tree. Support values at individual nodes for various combinations of alignment sets and methods are presented in accompanying tables. The posterior probabilities ≤0.5 and bootstrap percentages ≤50 were indicated by a dash “-”. The node 8 received no support under these thresholds
