Fig. 2.
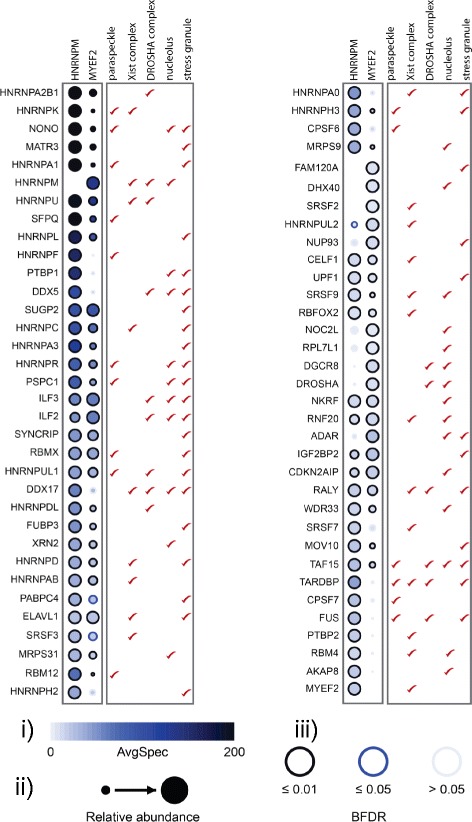
BioID of BirA*-FLAG-tagged HNRNPM and MYEF2 from HEK293 cells highlighting significant groups of preys annotated to be in specific RNA granules or RNA associated complexes. Prey proteins found in different RNA-associated compartments are based on published data: Paraspeckle [45], X chromosome inactivation complex [48], DROSHA microprocessor complex [50–52], Stress granule [53], and Nucleolus (using g:Profiler; http://biit.cs.ut.ee/gprofiler/index.cgi [111]. Data are the mean of two biological replicate experiments. (i) Spectral counts of each prey protein are displayed as a gradient of intensity in blue as indicated in the scale. (ii) Relative abundance between HNRNPM and MYEF2.(iii) Bayesian false discovery rate. See also Additional file 1: Tables S3 and S4
