Abstract
Background
Pituitary adenoma (PA) is a benign brain tumor that can cause neurological, endocrinological and ophthalmological aberrations. Till now there is a need to identify factors that can influence the tumor invasiveness and recurrence. The aim of this study was to evaluate the associations between the signal transducer and activator of transcription 3 (STAT3) promoter methylation, mRNA expression and the invasiveness or recurrence of PAs and patient clinical characteristics.
Methods
Study participants comprised of 102 subjects with a diagnosis of PA: 54 functioning and 48 non-functioning, 58 invasive and 30 non-invasive PAs and 14 relapses. The bisulfite treatment of tumor DNA and methylation-specific polymerase chain reaction (MS-PCR) method was used to determine the STAT3 gene promoter methylation. For the STAT3 mRNA expression, the first-strand cDNA was produced from total RNA by using reverse transcriptase and quantitative real-time PCR (qRT-PCR) was performed.
Results
In 10.78% (11/102) of PA tissues STAT3 gene promoter was methylated. A gender of male and patient group older than 60 years were significantly associated with reduced STAT3 mRNA expression (Mann-Whitney test, p = 0.025, p = 0.047, respectively). However, no more statistical differences were found between STAT3 promoter methylation, mRNA expression and patient clinical characteristics or PA invasiveness or recurrence.
Conclusions
Further investigations are needed to clarify the influence of STAT3 gene promoter methylation and mRNA expression changes in PAs.
Keywords: Pituitary adenoma, Signal transducer and activator of transcription 3, DNA methylation, mRNA expression, Invasiveness
Background
Pituitary adenomas (PA) arise from adenohypophyseal cells and account for 10–15% of all intracranial neoplasms. About 0.2% of all PAs with subarachnoid, brain or systemic metastasis are considered to be malignant [1]. Historically, PA tumors have been classified according to size as microadenomas (less than 1 cm) or macroadenomas (greater than or equal to 1 cm). Depending on PAs hormonal activity, they are also classified as either functional - secreting adrenocorticotropic hormone (ACTH), growth hormone (GH), prolactin (PRL), thyroid-stimulating hormone (TSH), luteinizing hormone (LH), and follicle-stimulating hormone (FSH) or non-functional - do not cause clinical hormone hypersecretion [2].
Surgical resection is the first-line of treatment for PAs, except for PRL-producing adenomas. Residual or recurrence tumors require re-operation, medical treatment or radiation [3]. However, the prediction of pituitary tumor behavior remains a challenge and factors known to be involved in pituitary tumorigenesis are not predictive of invasiveness or aggressiveness [4, 5]. Therefore, a better understanding of PA tumorigenesis is important for the development of novel targeted drugs.
In many cancers, including PAs, epigenetic changes have been involved in development of tumors [6]. Pituitary tumorigenesis frequently involves mutations of oncogenes (Cyclin D1, C-myc), tumor suppressor genes (MEN1, p53), cell cycle regulator genes (AIP, PRKAR1A), and epigenetic factors (DNA methylation, imprinting, histone modification) [2, 6]. For example, the methylation and deletion of DAP-kinase gene was associated with highly invasive and/or metastatic pituitary tumors [7]. Also, the methylation of the growth arrest and DNA damage-inducible gene (GADD45G) leads to gene silencing and is associated with the development of somatotropinomas, prolactinomas, and non-functioning adenomas [8].
One of the potential marker for PA diagnosis and prognosis could be signal transducer and activator of transcription 3 (STAT3) gene, a member of the STAT family of proteins that mediates cytokine signaling and nuclear transcription [9]. STAT3 forms dimers when activated by tyrosine kinase signals and translocates to the nucleus to regulate expression of genes by binding to elements within promoters [10]. In vitro and in mouse tumor models STAT3 is integrally involved in tumorigenesis, including apoptosis, cell cycle progression, tumor angiogenesis, invasion and metastasis [11–13]. However, there were no studies about STAT3 gene DNA and mRNA expression changes in invasive or recurrence PAs.
To this purpose, we performed methylation-specific polymerase chain reaction (MS-PCR) to evaluate methylation status of STAT3 gene promoter and real-time polymerase chain reaction (RT-PCR) to determine the mRNA expression changes in PAs. These epigenetic alterations were then correlated with clinical features of the patients and the invasiveness, recurrence of PAs.
Methods
The subject
The study was performed in the Neuroscience Institute, Lithuanian University of Health Sciences between 2010 and 2016. After surgical resection tumor tissues were frozen in nitrogen liquid and the following clinical characteristics were gathered: gender, age at the time of the operation, relapse, tumor size, PA activity, invasiveness, hypersecretion of PRL, GH, IGF-1, ACTH or more than one hormone and diagnoses of Cushing syndrome, acromegaly or prolactinoma.
Study participants comprised of 102 patients (56.86% women and 43.14% men, 60.78% older than 60 years and 39.22% younger than 60 years). Clinical findings showed that the endocrinological features were: 54 functioning and 48 non-functioning adenomas. Functioning adenomas were: 4 GH-secreting adenomas, 2 IGF-1-secreting adenomas, 36 PRL-secreting adenomas, 1 ACTH-secreting adenoma and 11 adenomas secreting more than one hormone. Also, magnetic resonance imaging findings and the Hardy classification, modified by Wilson was used to quantify the sphenoid sinus invasion and suprasellar extension [14]. In connection with the fact that not for all patients were able to receive data, relapse was established in 14 and invasiveness - in 88 patients. From them, it was found 58 invasive and 30 non-invasive PAs. It is important to mention that all PA tumors were macroadenomas.
Methylation-specific polymerase chain reaction (MS-PCR)
For the DNA methylation studies, 102 frozen PA tissues were used. First, DNA was extracted by 10% “UtraPure™ ”SDS (Invitrogen, USA), proteinase K (Carl Roth® GmbH, Germany), followed by phenol–chloroform (Carl Roth® GmbH, Germany) and 96% ethanol (Stumbras, Lithuania) precipitation. Subsequently, according to the manufacturer’s instructions, genomic DNA was modified with sodium bisulfite using EZ DNA methylation kit™ (Zymo Research, USA) and eluted in 40 μL of nuclease-free water.
The MS-PCR was performed in 15 μl of a mixture containing 10 pmol of each primer (Metabion International AG, Germany), 7.5 μL Maxima® Hot Start PCR Master Mix (ThermoFisher Scientific, USA) with Hot Start Taq DNA polymerase and nuclease-free water. MethPrimer online tool [15] was used to design primers for methylated STAT3 allele: 5′-TTTTGGGTGGTCGAACG-3′ (forward), 5′-AAAAACAACGCCAAACCG-3′ (reverse), resulting in a 222 bp PCR amplicon and for non-methylated allele: 5′-ATTTTTGGGTGGTTGAATG-3′ (forward), 5′-AAAAAAAACAACACCAAACC-3′ (reverse), resulting in a 225 bp PCR amplicon. The reaction was hot started at 95 °C for 5 min and MS-PCR conditions for all of the reactions were as follows: denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s and extension at 72 °C for 15 s, for 38 cycles, and final 5 min extension at 72 °C. In each set of methylation-specific PCR reactions three controls were included: normal human blood lymphocyte DNA treated with bisulfite (non-methylated control), the Bisulfite-Converted Universal Methylated Human DNA Standard (Zymo Research, USA) (methylated control) and nuclease-free water (negative control).
The MS-PCR products were analyzed on a 2% agarose gel with ethidium bromide (10 mg/ml, Invitrogen, USA) by horizontal electrophoresis.
Quantitative real-time PCR (qRT-PCR)
For the STAT3 gene mRNA expression analysis, total RNA was extracted from the same set of PA samples (102 samples) that has been used for STAT3 gene methylation analysis with Trizol reagent (Ambion, Life Technologies, USA), according to the manufacturer’s instructions and stored at −80 °C. RevertAid H Minus M-MuLV Reverse Transcriptase (ThermoFisher Scientific, USA) and random hexamer primers (ThermoFisher Scientific, USA) were used to produce the first-strand cDNA. Total reaction volume was 20 μl. Negative controls without reverse transcriptase were prepared as above.
Quantitative real-time PCR for STAT3 and β-actin was performed in a Real-Time PCR System “Applied Biosystems 7500 Fast”(Applied Biosystems, USA) with SYBR Green chemistry. The qRT-PCR was carried out in a 12 μl of mixture which consisted of 6 μl Maxima SYBR Green/ROX qPCR Master Mix (2X) (ThermoFisher Scientific, USA), 15 ng of the cDNA, nuclease-free water and STAT3 gene-specific primers that were designed according to the published data [16]: 5′-CATATGCGGCCAGCAAAGAA-3′ (forward), 5′-ATACCTGCTCTGAAGAAACT-3′ (reverse), resulting in a 152 bp PCR amplicon to a total concentration of 0.3 μM. The housekeeping gene β-actin with primers designed according to the published data [17]: 5′- AGAGCTACGAGCTGCCTGAC-3′ (forward), 5′-AGCACTGTGTTGGCGTACAG-3′ (reverse), resulting in a 184 bp PCR amplicon to a total concentration of 0.1 μM, was used as an internal control. The PCR amplification cycles were as follows: the denaturation step at 95 °C for 10 min and then 92 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s for 40 cycles and a final step for the generation of a melting curve. Also, GAPDH TaqMan assay (Applied Biosystems, Hs02758991_g1, cat. No. 4331182) was used to verify the expression of another endogenous control in PAs. The PCR amplification was performed according to the manufacturer’s protocol. All sample measurements were performed in triplicate. Healthy human brain RNA sample (RHB) “FirstChoice Human Brain Reference RNA” (Ambion, Life Technologies, USA, cat. No. AM6050) was used for standard curve design and the parameters were: for STAT3: efficiency 92.68%, R2 = 0.99, slope − 3.51; for β-actin: efficiency 100.08%, R2 = 0.99, slope − 3.32.
Finally, results were presented as 2-ΔΔCt calculations, where ΔΔCt = (Ct,STAT3 - Ct, internal controls)Sample x - (Ct,STAT3 - Ct, internal controls)RHB [18].
Statistical analysis
Statistical analyses were conducted with the SPSS Statistics 19 (SPSS Inc., Chicago, IL) and GraphPad Prism 7 (California, USA) software packages. The Mann-Whitney test was used to find the associations between STAT3 gene mRNA expression and the methylation status, also the clinical factors: gender, age, relapse, Cushing syndrome, acromegaly, prolactinoma, invasiveness, secreting and non-secreting PAs and hormone groups. The associations among STAT3 gene promoter methylation and clinical characteristics of PA patients were assessed with Chi-square test and p < 0.05 values were considered statistically significant.
Results
Analysis of STAT3 methylation frequency in patients with PA
STAT3 gene methylation status was performed in 102 PA samples by the MS-PCR method. The representative chart of MS-PCR products that were separated by horizontal electrophoresis and visualized under UV illumination is shown in Fig. 1. All PA samples were compared with positive and negative controls. Our results showed that STAT3 gene was methylated in 10.78% (11/102) and non-methylated in 89.22% (91/102) of PA samples. These results indicate that STAT3 has low methylation status in PAs.
Fig. 1.
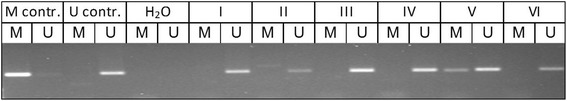
Analysis of STAT3 gene MS-PCR products on 2% agarose gel. M indicates methylated alleles, U unmethylated alleles. M cont. – positive methylation control (Standard Bisulfite Converted Universal Methylated Human DNA), U cont. – negative methylation control (normal human peripheral lymphocytes), H2O – water control, I-VI designate PA samples
Next, we tried to find out the associations between STAT3 gene promoter methylation and patient clinical characteristics: gender, age at the time of operation, relapse, PA function, invasiveness and diagnoses of Cushing syndrome, acromegaly or prolactinoma. To this purpose, Chi-square test was performed. However, statistical analysis showed no significant differences (Table 1). In addition, invasive PAs showed low methylation status of STAT3 gene. Only in 12.07% (7/58) invasive PAs STAT3 gene was methylated and non-methylated in 87.93% (51/58) cases but with no significance (χ2 test, p = 0.428).
Table 1.
Relationship between STAT3 promoter methylation, patient clinical characteristics and PA invasiveness
| STAT3 gene methylation | ||||
|---|---|---|---|---|
| Number of patients | M (%) | U (%) | p-value | |
| Cases | 102 | 11(10.78) | 91(89.22) | |
| Age (years) | ||||
| ≤ 60 | 40 | 4(10.00) | 36(90.00) | 0.837 |
| > 60 | 62 | 7(11.29) | 55(88.71) | |
| Gender | ||||
| Female | 58 | 6(10.34) | 52(89.66) | 0.870 |
| Male | 44 | 5(11.36) | 39(88.64) | |
| PA function | ||||
| Secreting | 54 | 6(11.11) | 48(88.89) | 0.910 |
| Non-secreting | 48 | 5(10.42) | 43(89.58) | |
| Relapse | ||||
| Appear | 8 | 1(12.50) | 7(87.50) | 0.871 |
| None | 94 | 10(10.64) | 84(89.36) | |
| Prolactinoma | ||||
| Appear | 36 | 4(11.11) | 32(88.89) | 1.000 |
| None | 18 | 2(11.11) | 16(88.89) | |
| Acromegaly | ||||
| Appear | 9 | 1(11.11) | 8(88.89) | 1.000 |
| None | 45 | 5(11.11) | 40(88.89) | |
| Cushing syndrome | ||||
| Appear | 1 | 0(0.00) | 1(100.00) | 0.721 |
| None | 53 | 6(11.32) | 47(88.68) | |
| Hormones | ||||
| PRL | 36 | 4(11.11) | 32(88.89) | |
| IGF-1 | 2 | 0(0.00) | 2(100.00) | |
| GH | 4 | 2 (50.00) | 2(50.00) | |
| ACTH | 1 | 0(0.00) | 1(100.00) | |
| Multiple | 11 | 0(0.00) | 11(100.00) | |
| Invasiveness | ||||
| Invasive | 58 | 7(12.07) | 51(87.93) | 0.428 |
| Non-invasive | 30 | 2(6.67) | 28(93.33) | |
M methylated, U non-methylated, PRL prolactin, IGF-1 insulin-like grow factor 1, GH growth hormone, ACTH – adrenocorticotropic hormone, multiple – PAs secreting more than one hormone
To further analysis we wanted to find out in which hormonally active PAs STAT3 gene promoter might be strongly methylated. Thus, the hormone distribution of PAs in methylated and non-methylated STAT3 gene promoter groups was performed. The analysis showed that with methylated STAT3 promoter, the quantities of PRL (4/36), IGF-1 (0/2), ACTH (0/1) and more than one hormone secreting PAs (0/11) hypersecretion were highly reduced as well as the non-secreting PAs (5/48) (Fig. 2). Nevertheless, the amount of GH hypersecretion was the same in methylated and non-methylated gene groups (2/4). However, the results were not statistically significant (Table 1).
Fig. 2.
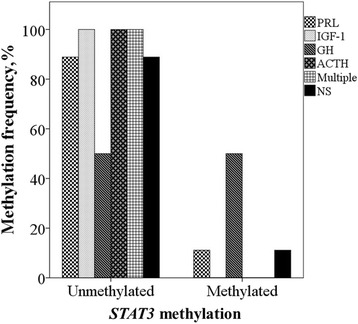
PA hormone distribution in methylated and non-methylated STAT3 gene groups. PRL – prolactin, IGF-1 – insulin-like grow factor 1, GH – growth hormone, ACTH – adrenocorticotropic hormone, multiple – PAs secreting more than one hormone, NS – non-secreting PAs
Analysis of STAT3 mRNA expression in patients with PA
To investigate the STAT3 mRNA expression changes in PAs, the qRT-PCR method was used. The values were normalized with two internal controls: GAPDH and β-actin. STAT3 mRNA expression was determined in 102 PA samples. It was found that in 13 cases there were no STAT3 mRNA expression. To compare the STAT3 mRNA expression in methylated and non-methylated gene groups, Mann-Whitney test was used. The analysis showed no significant differences of STAT3 mRNA expression between the group of methylated STAT3 (10/89 tumors) and the group of non-methylated STAT3 gene (79/89 tumors, p = 0.795).
Next, we assessed the associations between STAT3 mRNA expression and patient clinical characteristics. We found that gene mRNA expression was lower with males (36/89 cases) than females (53/89 cases, Mann-Whitney test, p = 0.025) and with patients older than 60 years (54/89 cases, Mann-Whitney test, p = 0.047, Fig. 3). However, there were no statistical differences with presence or absence of repeated surgery, invasiveness, secreting or non-secreting PAs, the diagnosis of acromegaly, prolactinoma or Cushing syndrome (Mann-Whitney test, p = 0.272, p = 0.798, p = 0.812, p = 0.935, p = 0.312 and p = 0.400, respectively).
Fig. 3.
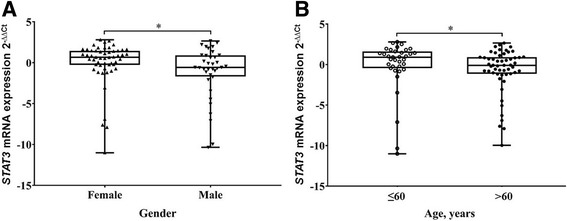
STAT3 mRNA expression associations with patient characteristics. a Box plots of STAT3 mRNA expression association with gender. b Box plots of STAT3 mRNA expression association with patient age. The line inside box represents the median, the lower and upper edges of the boxes represent the 25th and 75th percentiles, respectively, and upper and lower lines outside the boxes represent minimum and maximum values (error bars). The Mann-Whitney test showed, that males and patient group older than 60 years were significantly associated with reduced STAT3 mRNA expression (p < 0.05)
For further investigation STAT3 gene mRNA expression values were categorized into four levels. Values 0.5-fold lower than the gene mRNA expression median were indicated as “low” expression level (N = 37), expression values 0.5-fold higher - “high” level (N = 33), values ranging in between “low” and “high” were referred to as “medium” mRNA expression level (N = 19). In 13 samples there were no STAT3 mRNA expression. Then we analyzed PRL, IGF-1, GH, ACTH, more than one hormone and non-secreting PAs distribution in these expression levels. As was shown in Fig. 4, in most cases the hypersecretion of PRL (13/36), ACTH hormone (1/1) and the highest amount of non-secreting PAs (19/48) were determined at “low”STAT3 mRNA expression level (χ2 test, p = 0.321, p = 0.565 and p = 0.417, respectively). Similarly, GH (2/4) and more than one hormone hypersecretion (6/11) - at “high”STAT3 mRNA expression level (χ2 test, p = 0.758 and p = 0.493, respectively). The IGF-1 hormone hypersecretion (1/2) mostly occured at “high”STAT3 mRNA expression level and with no gene expression (χ2 test, p = 0.473).
Fig. 4.
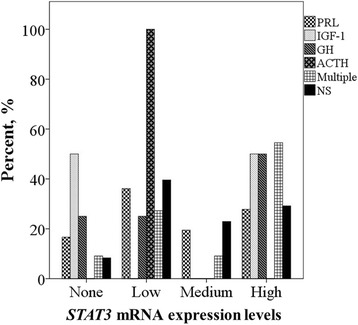
PA hormone distribution in STAT3 mRNA expression levels. PRL – prolactin, IGF-1 – insulin-like grow factor 1, GH – growth hormone, ACTH – adrenocorticotropic hormone, multiple – PAs secreting more than one hormone, NS – non-secreting PAs
In our last series of result analysis, we took the biggest group of hormonally active PAs (PRL-secreting PAs) and non-secreting PAs and asked if STAT3 mRNA expression is associated with PA invasiveness (Fig. 5). However, Mann-Whitney test did not reveal significant differences in STAT3 mRNA expression median values (p = 0.769 and p = 0.753, respectively).
Fig. 5.
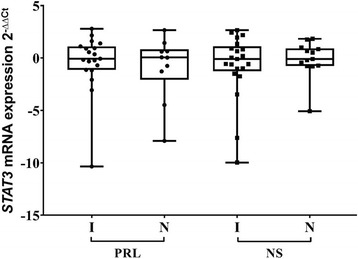
STAT3 mRNA expression changes in invasive and non-invasive PRL secreting and non-secreting PAs. The horizontal bars represent the mean values with 95% confidence interval, the spots represent the amount of samples. PRL – prolactin, NS – non-secreting PAs
Discussion
Pituitary adenoma is a benign brain tumor that could invade the sphenoid, cavernous sinus, or dura mater and some of them are described as aggressive, with a high proliferation rate and short postoperative time to recurrence [1, 19]. Therefore, an epigenetic marker to predict a greater malignant potential of PAs still remains to be established. STAT3 gene may be a promising target because it is aberrantly activated in a wide variety of human cancers and plays crucial roles in tumor cell proliferation, survival, invasion and tumor-promoting inflammation [20–26].
We conducted this study to examine the prognostic significance of STAT3 gene promoter methylation and mRNA expression in 102 PA patients and to determine the associations with patient clinical data. We determined 10.78% (11/102) STAT3 promoter methylation frequency. Also, in our study invasive PAs showed low STAT3 methylation status. Only in 12.07% (7/58) invasive PAs, STAT3 promoter was methylated (Table 1). We also demonstrated that PRL, IGF-1, ACTH and more than one hormone hypersecretion mostly occur with non-methylated STAT3 gene (Fig. 2). Low STAT3 gene methylation status indicates that this gene is little suppressed in PAs and might be constantly activated. However, mechanisms related to this are still unknown. Numerous studies showed the activation of STAT3 in a wide variety of tumors [20–26] and the proposed role for STAT3 in tumor formation or progression is based on the link between its activation and transformation [10]. For example, Morikawa T et al. (2011) found that individuals with STAT3-activated colorectal cancers experience a poorer prognosis, suggesting a potential tumor-promoting role of p-STAT3 expression as a prognostic biomarker [20]. Increased expression of phosphorylated STAT3 and JAK/STAT activation in glioma samples has also been correlated with significantly shorter overall survival and has been associated with more aggressive tumors, indicating that STAT3 is a key contributor to glioma pathogenesis by mediating cell survival, growth, and proliferation [21–23]. Moreover, in breast carcinoma cells, the STAT3 activation has been correlated with cell proliferation [24], in colon cancer STAT3 has been shown to be overexpressed [25], as well as in high-grade prostate cancer patients [26]. In these mentioned studies, STAT3 has been described as an oncogene, however, there are some other publications in which STAT3 has been shown to be a tumor suppressor. For example, in colorectal carcinoma biopsies STAT3 overexpression has been associated with an improvement in median survival of about 30 months [27]. French patient cohort by Monnien et al. (2010) provided evidence for a association of phosphorylated STAT3 appearance with prolonged survival of rectal cancer patients [28]. Therefore, STAT3 can act as an oncogene or a tumor suppressor, depending on tumor tissue. In our results, there were no other associations between STAT3 gene promoter methylation, mRNA expression and PA invasiveness, recurrence or patient clinical characteristics. Only a gender of male and patient group older than 60 years were significantly associated with reduced STAT3 mRNA expression (Mann-Whitney test, p = 0.025, p = 0.047, respectively). However, the mechanism of STAT3 expression changes in PA proliferation and invasion has not been reported yet, making it necessary to further research on the pro- and anti-tumorigenic effects of STAT3 gene activation.
It is important to mention, that Zhou et al. (2015) made a research of STAT3 expression in somatotroph adenomas. They found that STAT3 expression was significantly enhanced in somatotroph adenomas (67% ± 5%) as compared with non-secreting pituitary tumor expression (unpaired t test, P < 0.001) [29]. Also, they showed that GH induces somatotroph cell STAT3 phosphorylation and nuclear translocation. Based on these results, Zhou et al. (2015) postulated that GH participates in a positive autocrine or paracrine feedback for STAT3 induction and supported the hypothesis that blocking STAT3 suppresses somatotroph tumor growth and inhibits GH hypersecretion [29]. Our results also showed a tendency that the highest amount of GH was determined at “high” STAT3 mRNA expression level (Fig. 4), making it necessary for further investigation.
Conclusions
In conclusion, this is the first study that has demonstrated the STAT3 gene promoter methylation and mRNA expression in 102 PA samples and analyzed the relationships between STAT3 gene epigenetic changes and patient clinical characteristics: age at the time of operation, gender, diagnoses of prolactinoma, acromegaly or Cushing syndrome, PA activity, recurrence and invasiveness. Our data have revealed that STAT3 gene mRNA expression is significantly lower with male gender and patients older than 60 years. However, no more significant results were found. To this purpose, our designed STAT3 gene promoter methylation analysis and mRNA expression cannot be considered as a prognostic marker in PAs or further examination including new methylation sites investigation for STAT3 methylation-specific PCR is required to estimate the influence of epigenetic changes in PAs.
Acknowledgements
We kindly thank B. Glebauskienė (Department of Ophthalmology, Lithuanian University of Health Sciences, Kaunas, Lithuania) for help to gather patient hormonal activities and invasiveness.
Funding
This research was funded by LUHS Faculty of Medicine fond.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACTH
Adrenocorticotropic hormone
- GH
Growth hormone
- IGF-1
Insulin-like growth factor 1
- MS-PCR
Methylation-specific polymerase chain reaction
- PA
Pituitary adenoma
- PRL
Prolactin
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RHB
Human brain reference RNA
- STAT3
Signal transducer and activator of transcription 3
Authors’ contributions
IV and PV planned the experiments, AT gathered pituitary adenoma samples and patient data, IV performed the experiments, PV, DS and GS analyzed and interpreted data, IV and PV wrote the manuscript. All authors read and approved the article.
Ethics approval and consent to participate
The study was approved by Kaunas Regional Biomedical Research Ethics Committee (Number P2–9/2003) and written permission under the approval of Ethics Committee of Lithuanian University of Health Sciences was taken for every patient.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Indre Valiulyte, Phone: +37064546134, Email: Indre.Valiulyte@lsmuni.lt.
Giedrius Steponaitis, Email: giedrius.steponaitis@lsmuni.lt.
Daina Skiriute, Email: daina.skiriute@lsmuni.lt.
Arimantas Tamasauskas, Email: a.tamasauskas@lsmuni.lt.
Paulina Vaitkiene, Email: paulina.vaitkiene@lsmuni.lt.
References
- 1.Kaltsas GA, Nomikos P, Kontogeorgos G, Buchfelder M, Grossman AB. Clinical review: diagnosis and management of pituitary carcinomas. J Clin Endocrinol Metab. 2005;90(5):3089–3099. doi: 10.1210/jc.2004-2231. [DOI] [PubMed] [Google Scholar]
- 2.Kalra RR, Taussky P, Niazi T, Couldwell W. Pituitary tumors: genetics and heritable predisposition. Tumors of the central nervous system. 2013;10:71–83. [Google Scholar]
- 3.Fukuoka H, Takahashi Y. The role of genetic and epigenetic changes in pituitary tumorigenesis. Neurol Med Chir. 2014;54:943–957. doi: 10.2176/nmc.ra.2014-0184. [DOI] [PubMed] [Google Scholar]
- 4.Al-Shraim M, Asa SL. The 2004 World Health Organization classification of pituitary tumors: what is new? Acta Neuropathol. 2006;111:1–7. doi: 10.1007/s00401-005-1093-6. [DOI] [PubMed] [Google Scholar]
- 5.Figarella-Branger D, Trouillas J. The new WHO classification of human pituitary tumors: comments. Acta Neuropathol. 2006;111:71–72. doi: 10.1007/s00401-005-1099-0. [DOI] [PubMed] [Google Scholar]
- 6.Tichomirowa MA, Daly AF, Beckers A. Familial pituitary adenomas. J Intern Med. 2009;266:5–18. doi: 10.1111/j.1365-2796.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 7.Simpson DJ, Clayton RN, Farrell WE. Preferential loss of death associated protein kinase expression in invasive pituitary tumours is associated with either CpG island methylation or homozygous deletion. Oncogene. 2002;21(8):1217–1224. doi: 10.1038/sj.onc.1205195. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Sun H, Danila DC, Johnson SR, Zhou Y, Swearingen B, Klibanski A. Loss of expression of GADD45 gamma, a growth inhibitory gene, in human pituitary adenomas: implications for tumorigenesis. J Clin Endocrinol Metab. 2002;87(3):1262–1267. doi: 10.1210/jcem.87.3.8315. [DOI] [PubMed] [Google Scholar]
- 9.Kim JE, Patel M, Ruzevick J, Jackson CM, Lim M. STAT3 activation in Glioblastoma: biochemical and therapeutic implications. Cancers. 2014;6:376–395. doi: 10.3390/cancers6010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 11.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers. 2014;6:926–957. doi: 10.3390/cancers6020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Jove R. The STATs of cancer - new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 13.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 14.Wilson CB. Neurosurgical management of large and invasive pituitary tumors. In: Tindall GT, Collins WF, editors. Clinical Management of Pituitary Disorders. New York: New York Raven Press; 1979. pp. 335–342. [Google Scholar]
- 15.MethPrimer. http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi. Accessed 13 Sept 2013.
- 16.Falzacappa MVV, Spasic MV, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109(10):353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 17.Skiriutė D, Steponaitis G, Vaitkienė P, Mikučiūnas M, Skauminas K, Tamašauskas A, Kazlauskas A. Glioma malignancy-dependent NDRG2 gene Methylation and Downregulation correlates with poor patient outcome. J Cancer. 2014;5:446–456. doi: 10.7150/jca.9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using RealTime quantitative PCR and the 2-∆∆ct method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Meij BP, Lopes MB, Ellegala DB, Alden TD, Laws ER. The long-term significance of microscopic dural invasion in 354 patients with pituitary adenomas treated with transsphenoidal surgery. J Neurosurg. 2002;96:195–208. doi: 10.3171/jns.2002.96.2.0195. [DOI] [PubMed] [Google Scholar]
- 20.Morikawa T, Baba Y, Yamauchi M, Kuchiba A, Nosho K, Shima K, Tanaka N, Huttenhower C, Frank DA, Fuchs CS, Ogino S. STAT3 expression, molecular features, inflammation patterns and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17(6):1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neuro-Oncol. 2010;100:339–343. doi: 10.1007/s11060-010-0195-8. [DOI] [PubMed] [Google Scholar]
- 22.Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong LY, Fuller GN, Hiraoka N, Priebe W, Sawaya R, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu Y, Zhong Y, Fu J, Cao Y, Fu G, Tian X, Wang B. Activation of JAK/STAT signal pathway predicts poor prognosis of patients with gliomas. Med Oncol. 2011;28:15–23. doi: 10.1007/s12032-010-9435-1. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Shaw PE. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J Biol Chem. 2002;277:17397–17405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- 25.Klampfer L. The role of signal transducers and activators of transcription in colon cancer. Front Biosci. 2008;13:2888–2899. doi: 10.2741/2893. [DOI] [PubMed] [Google Scholar]
- 26.Kroon P, Berry PA, Stower MJ, Rodrigues G, Mann VM, Simms M, Bhasin D, Chettiar S, Li C, Li PK, et al. JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res. 2013;73(16):5288–5298. doi: 10.1158/0008-5472.CAN-13-0874. [DOI] [PubMed] [Google Scholar]
- 27.Gordziel C, Bratsch J, Moriggl R, Kno T, Friedrich K. Both STAT1 and STAT3 are favourable prognostic determinants in colorectal carcinoma. Br J Cancer. 2013;109:138–146. doi: 10.1038/bjc.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monnien F, Zaki H, Borg C, Mougin C, Bosset JF, Mercier M, Arbez-Gindre F, Kantelip B. Prognostic value of phosphorylated STAT3 in advanced rectal cancer: a study from 104 French patients included in the EORTC 22921 trial. J Clin Pathol. 2010;63:873–878. doi: 10.1136/jcp.2010.076414. [DOI] [PubMed] [Google Scholar]
- 29.Zhou C, Jiao Y, Wang R, Ren AG, Wawrowsky K, Melmed S. STAT3 upregulation in pituitary somatotroph adenomas induces growth hormone hypersecretion. J Clin Invest. 2015;125(4):1692–1702. doi: 10.1172/JCI78173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


