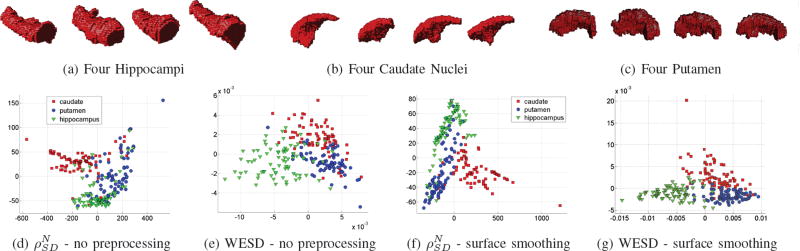
Fig. 8.
2D embedding of subcortical structures: 240 structures (80 caudate nucleus, 80 putamen and 80 hippocampus) are extracted from MR scans of 40 different individuals. (a),(b) and (c) show some example structures from this dataset. Note the high intra-class variability and the artefacts due to finite resolution and manual segmentations. (d) and (e) plot 2D embeddings of these 240 structures obtained based on the affinity matrices computed via and WESD respectively. These embeddings are computed without any preprocessing applied to the structures. The embedding obtained with WESD distinctly clusters the objects with respect to the anatomical structures. The embedding in (d) however, shows some ambiguities in the separation. Graphs in (f) and (g) plot the similar embeddings obtained after smoothing the surfaces of the structures to remove artefacts. The embedding obtained by , although better than (d), still suffer from similar problems. The embedding based on WESD on the other hand, now even between better separates the groups.