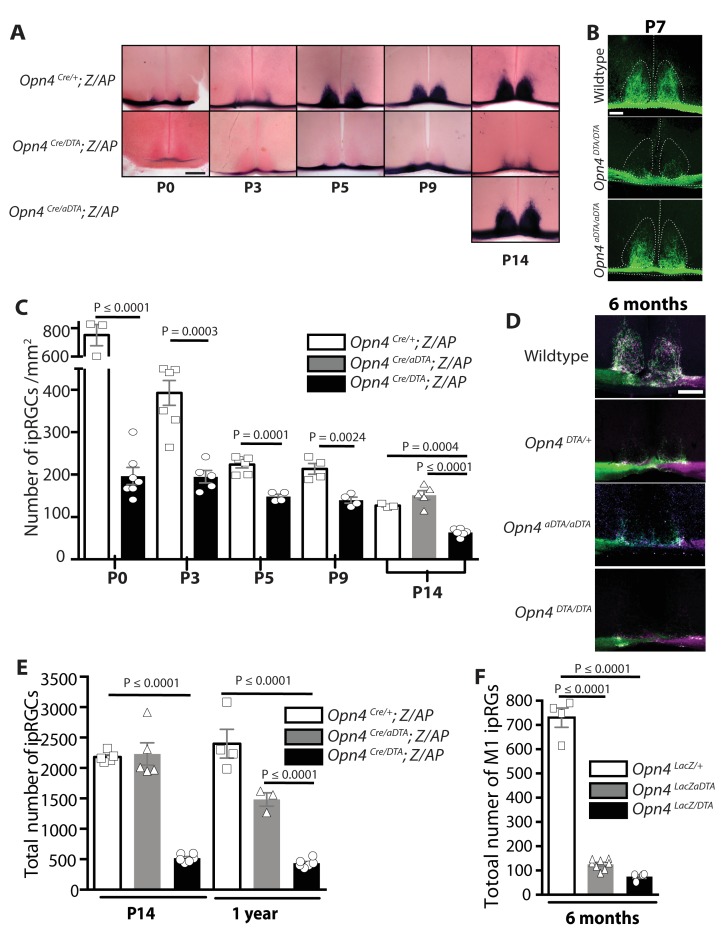
Figure 1. Developmental ablation of ipRGCs in the mouse retina.
(A) Developmental time course of ipRGC innervation of the SCN, visualized by AP staining, in Opn4Cre/+; Z/AP and Opn4Cre/DTA; Z/AP mouse. For comparison, SCN staining from Opn4Cre/aDTA; Z/AP mice at P14 are also shown. Scale bar = 200 μm. (B) SCN innervation in P7 WT, Opn4DTA/DTA, and Opn4aDTA/aDTA mice revealed by CTB injections into the eyes. Scale bar = 100 μm. (C) Developmental time course of ipRGC (all subtypes) cell density visualized by AP staining of retina from Opn4Cre/+ Z/AP (control) and Opn4Cre/DTA; Z/AP mice at P0 (control n = 3, DTA n = 7), P3 (control n = 7, DTA n = 5), P5 (control n = 6, DTA n = 4), P9 (control n = 4, DTA n = 4), and P14 (control n = 3, DTA n = 6, aDTA n = 5). Cell counts from P14 retinas of Opn4Cre/aDTA; Z/AP mice are also shown for comparison. Using a two-way ANOVA, we found a strongly significant effect of genotype. A t-test for P0, P3, P5, and P9 time points, and a one-way ANOVA with Bonferroni's post-hoc analysis for P14 revealed a significant cell loss at each time point. (D) SCN innervation revealed by CTB injections into the eyes of 6-month-old WT, Opn4DTA/+, Opn4DTA/DTA, and Opn4aDTA/aDTA mice. Scale bar = 200 μm. (E) Total cell counts of ipRGCs (all subtypes) revealed by alkaline phosphatase staining at P14 and 1 year of age in Opn4Cre/+; Z/AP (control; P14: n = 5, 1 year: n = 4), Opn4Cre/aDTA; Z/AP (P14: n = 5; 1 year: n = 3), and Opn4Cre/DTA; Z/AP (P14: n = 6; 1 year: n = 6). Two-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. (F) Total cell counts of M1 ipRGCs, identified by x-Gal staining of retinas from 6 month old Opn4LacZ/+ (control; n = 4), Opn4LacZ/aDTA, and Opn4LacZ/DTA mice (n = 4). One-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. Error bars represent s.e.m. for all graphs. See also Figure 1—figure supplement 1.