Figure 4. The GET and SND pathways act as backup for targeting in-vivo.
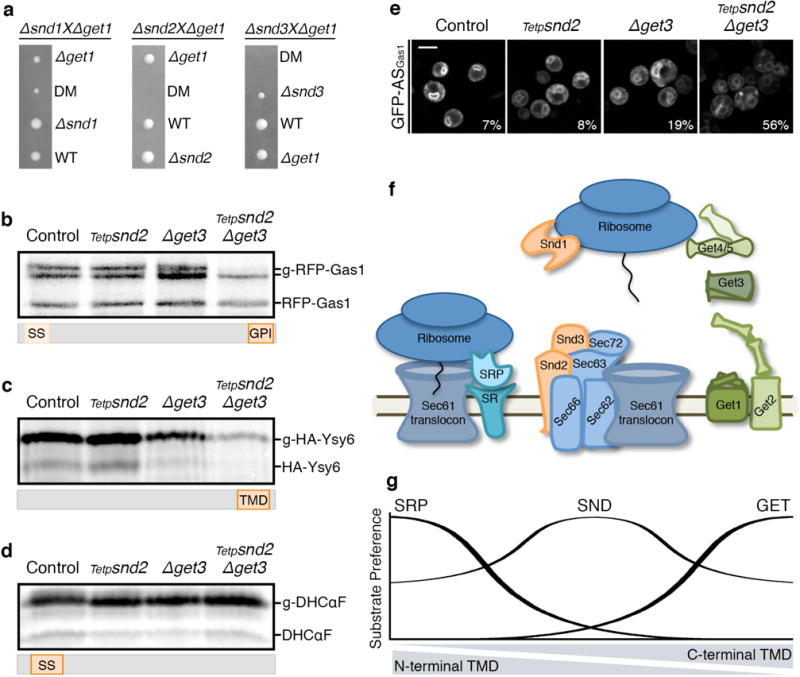
(a) Tetrads from Δsnd/Δget diploids demonstrate a synthetic sick/lethal interaction.
(b–d) Metabolic labeling of RFP-Gas1 (b), HA-Ysy6 (c) or DHCαF (d) showing decrease in translocated forms only for SRP-independent substrates in the conditional SND2/GET3 double-mutant. Accumulation of pre-inserted forms cannot be observed due to lack of proteasomal inhibition. Results repeated in three biological replicates.
(e) GFP fused to Gas1 GPI-anchoring sequence (GFP-ASGas1). Percentage of cells (from 100) with mistargeting depicted on images. Scale bar, 5 μm.
(f) Scheme of the eukaryotic ER-targeting apparatus.
(g) Model of the ER-targeting pathways’ interplay.
For gel source data see Supplementary Figure 1.
