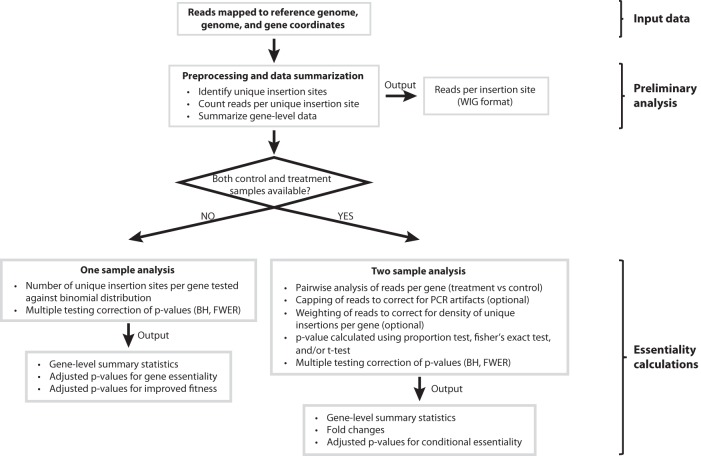
FIG 1 .
Workflow for Tn-seq analysis using TSAS. Genome sequence (FASTA format), gene coordinates (GFF v3 format), and mapped DNA sequencing reads (reads) (using Bowtie [version 1 or 2], SOAP, or ELAND) are provided as input to TSAS for essentiality analysis. After preliminary analysis to identify transposon insertion sites and count reads mapping to those sites, a one- or two-sample analysis can be performed depending on sequencing data available. See the TSAS User Guide (Text S1) for details regarding one- and two-sample analysis settings, options, and outputs. BH, Benjamini-Hochberg; FWER, family-wise error rate.