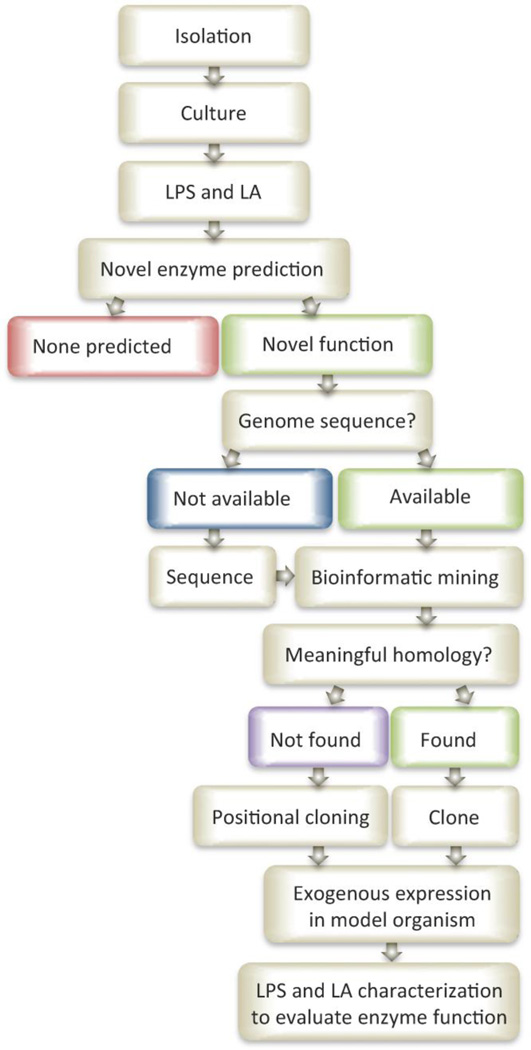
Figure 3. Pathway to systematic identification of novel lipid A biosynthetic and/or modifying enzymes.
Species from exotic source libraries (marine, thermal vents, etc.) must first be isolated and cultured for LPS and lipid A extraction. By comparing the known lipid A structures and their respective biosynthetic pathways to novel structures identified from exotic sources predictions can be made about new, unique, or alternative enzymatic functions. Where novel enzymes are predicted, homology searches of the available genome(s) can be performed using similar enzymes of known function and sequence. Some species may require genome sequencing and assembly prior to bioinformatic mining. Homology searches resulting in meaningful predictions can be cloned and exogenously expressed in a model organism and further evaluated at the lipid A structural modification level. Homology searches not yielding meaningful predictions may lead to gene identifications using a more laborious positional cloning approach for the most interesting potential modifying enzymes. Careful consideration of atypical growth conditions, unique inducing conditions, and unforeseen technical hurdles will be necessary.