Abstract
The high and low alcohol-drinking (HAD and LAD) rats were selectively bred for differences in alcohol intake. The HAD/LAD rats originated from the N/Nih heterogeneous stock (HS) developed from intercrossing 8 inbred rat strains. The HAD × LAD F2 were genotyped, and a powerful analytical approach, utilizing ancestral recombination as well as F2 recombination, was used to narrow a QTL for alcohol drinking to a 2 cM region on distal chromosome 10 that was in common in the HAD1/LAD1 and HAD2/LAD2 analyses. Quantitative real time PCR (qRT-PCR) was used to examine mRNA expression of 6 candidate genes (Crebbp, Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3) located within the narrowed QTL region in the HAD1/LAD1 rats. Expression was examined in 5 brain regions, including the nucleus accumbens, amygdala, caudate putamen, hippocampus, and pre-frontal cortex. All six genes showed differential expression in at least one brain region. Of the genes tested in this study, Crebbp and Mapk8ip3 may be the most promising candidates with regard to alcohol drinking.
Keywords: HAD and LAD rats, QTL, fine mapping, gene expression
Introduction
The impact of genes on the risk for developing alcoholism has been firmly established in humans. There is also a strong genetic component in the response to alcohol in animals. Animal models with phenotypic traits similar to the human condition have been useful in identifying genetic loci that may influence this complex disease. One animal model, the High- and Low-Alcohol Drinking (HAD and LAD) rats, has been used to map quantitative trait loci (QTL) for alcohol preference and consumption. The HAD1/LAD1 and the HAD2/LAD2 rats were independently derived from the N/Nih heterogeneous stock rats, developed by intercrossing 8 inbred rat strains (ACI/N, WKY/N, F344/N, BUF/N, BN/Ssn, WN/N, M520/N, and Mr/N) with a wide range of alcohol preference and other alcohol-related phenotypes (Hansen & Spuhler, 1984; Li & Lumeng, 1984; Li et al., 1993; Spuhler & Deitrich, 1984). Consequently, both the HAD1/LAD1 and HAD2/LAD2 rat lines are genetically diverse, and theoretically, could have as many as 8 alleles at any given loci in which all parent strains differ.
In a previous study, a QTL for alcohol preference was identified on distal chromosome 10 in the HAD1/LAD1 rats (Foroud et al., 2000) and later confirmed in the HAD2/LAD2 lines (Carr et al., 2003). Because the QTL on chromosome 10 exhibited linkage in both studies, further assessment of the region was warranted. Therefore, in the present study, we used a powerful analytical approach, utilizing ancestral recombination (from the aforementioned 8 inbred strains) as well as F2 recombination to narrow the chromosome 10 QTL in both the HAD1/LAD1 and HAD2/LAD2 lines. This approach is implemented computationally by calculating the probability that a marker genotype or haplotype is inherited identical by descent (IBD) from each of the 8 inbred founders crossed to generate the N/Nih HS and subsequently the HAD × LAD F2 animals. This in turn allows the QTL to be mapped to a narrower segment among the F2 progeny with more extreme phenotypic values. This study design and analytic approach was successfully implemented in a recent study using HS mice (Valdar et al., 2006).
To further assess the molecular mechanisms underlying this QTL, we took advantage of unpublished microarray data that were available for the iHAD1/iLAD1 rats, which are inbred strains developed from the HAD1/LAD1 lines. We examined mRNA expression of six genes in a 2 cM QTL region that was in common in the HAD1/LAD1 and HAD2/LAD2 analysis. Five genes within this region (Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3) were differentially expressed in microarray (W.J. McBride, personal communication). We also examined the Crebbp gene, which falls within the region and has been shown to play an important role in alcohol addiction. qRT-PCR was used to examine expression differences for each of the 6 genes in five brain regions of the HAD1/LAD1 rats, including the nucleus accumbens, amygdala, caudate putamen, hippocampus, and pre-frontal cortex.
Materials and Methods
Subjects used for genotyping
Previously, six reciprocal crosses of HAD1 and LAD1 rats taken from the 26th generation of selective breeding were completed (Foroud et al., 2000). Six F1 progeny from each cross (3 females and 3 males) were intercrossed 2 to 3 times to create 459 HAD1 × LAD1 F2 animals. Subsequently, six reciprocal crosses of HAD2 and LAD2 rats taken from the 36th generation were performed. Six F1 progeny of each cross (4 females and 2 males) were then intercrossed 2 to 3 times to create 428 HAD2 × LAD2 F2 animals. In addition, the current dataset also included one DNA sample from each of the 8 inbred ancestral strains that were used in this study.
Phenotyping of HAD × LAD F2 Rats
The method for phenotyping was identical in the HAD1/LAD1 and HAD2/LAD2 lines. Each animal was housed individually and given free choice of water and a 10% (v/v) ethanol solution. Food was available ad libitum throughout the testing period to ensure that consumption was not for caloric intake. The volume of ethanol consumed was recorded every 3 days for a period of 3 weeks. The location of the bottles was switched each time to control for position bias. All nine scores were averaged to obtain an alcohol consumption score. Drinking scores were also used to calculate alcohol preference, which is the volume of 10% ethanol divided by the total fluid volume consumed. Because drinking scores from HAD1/LAD1 are quite different from HAD2/LAD2, normal quantiles were used in the analyses.
DNA Isolation and Genotyping
Genomic DNA was isolated from the spleens of each HAD1/LAD1 animal by using Purgene (Gentra Systems, Inc. Minneapolis, MN) according to the manufacturer’s protocol, and tail snips of each HAD2/LAD2 animals using GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, Inc. St. Louis, MO). For further details on the DNA isolation procedures, see Carr et al. (2003).
Most of the microsatellite markers analyzed in this study were genotyped formerly in the HAD1/LAD1 and HAD2/LAD2 lines (see Carr et al., 1998; Carr et al., 2003) for marker selection and genotyping procedures). Not all markers were informative in both crosses. Four additional markers (D10rat183, D10rat121, D10rat17, and D10rat49) were added to chromosome 10 in the HAD2/LAD2 animals to increase the marker density within the QTL region. In addition, the 8 ancestral inbred strains were genotyped for each marker used in this study. When additional genotyping was performed, the procedures were identical to those found in Carr et al. (1998).
Genetic analysis
QTL analyses was performed by using a software package “HAPPY” (Mott et al., 2000; http://www.well.ox.ac.uk/happy). At the first stage, a dynamic-programming algorithm is used to calculate FLi(s,t), which is the probability of a given marker interval L descended from founder strains s and t for each individual. Then a regress model was fitted to test the presence of QTL. TL (s,t) is the phenotypic effect contributed by strains s, t at marker interval L. If T(s,t) = T(s) +T(t), then it is an additive model. When interactions are included, it is defined as a full model. The QTL interval was defined by that region in which the LOD score was above 3.0. The mapping approach results in a stepwise LOD score so the region was defined based on the position of the marker at which the LOD score fell below 3.0. This results in a conservative narrowing of the region.
Animals and Dissection of Brain Regions
Inbred adult male rats, 90–100 days old, from the iHAD strains 2A (n = 4) and 3A (n = 2) and iLAD strains 7A (n = 2) and 5C (n = 4) were used in the microarray experiments. They were all from the 25th generation. Inbreeding by brother-sister mating was initiated after the S30 generation of mass selection and was in the F37 generation at the start of these experiments. For the qRT-PCR study, adult selectively bred male HAD1 and LAD1 rats (90–100 days old) were used in the experiments. All animals (both inbred and selectively bred) were received in our facilities 3 weeks prior to the experiment. Rats were double housed on a 12:12 light dark cycle with lights on at 0700 hours. Rats had water and rat chow ad libitum. Animals were habituated to handling and to the guillotine daily between 0900 – 1000 hr for 10 days prior to sacrifice. The animals used in these experiments were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All research protocols were approved by the Institutional Animal Care and Use Committee and are in accordance with the guidelines of the Institutional Animal Care and Use Committee of the National Institute on Drug Abuse, NIH, and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council 1996).
Animals were sacrificed by decapitation between 0900 and 1000 hr over consecutive days, with equal numbers of animals from each strain sacrificed each day. This minimized differences in time of sacrifice and dissection, and maintained the experimental balance across the two strains. The head was immediately placed in a cold box maintained at −15°C, where the brain was rapidly removed and placed on a glass plate for dissection. All equipment used to obtain tissue was treated with RNAse Zap (Ambion, Inc. Austin, TX) to prevent RNA degradation. The nucleus accumbens, pre-frontal cortex, amygdala, hippocampus, and caudate putamen were dissected bilaterally according to the coordinates of Paxinos and Watson (1998). These subregions were selected because they have been implicated in alcohol-drinking behavior (McBride and Li, 1998). The nucleus accumbens and caudate putamen are dissected from a 2 mm section generated by a coronal cut at 2 mm anterior to the optic chiasm (Bregma 1.70 mm) and a coronal cut at the optic chiasm (Bregma 0.26 mm). The nucleus accumbens is dissected bilaterally by cutting below the rhinal fissure and trimming off the olfactory tubercles and cortical tissue at the ventral and ventrolateral borders of the slice. From the remaining tissue the caudate putamen (bilateral) is dissected below the corpus callosum leaving the septum hanging in the center. The amygdala is dissected by a cut at the lateral borders of the lateral hypothalamus (Bregma 2.12 mm) and ventral of the rhinal fissure, with cortical tissue then removed at the lateral edges of the dissected slice. The caudal border of the amygdalar dissection is the rostro-ventral border of the CA3 subfield of Ammon’s horn (4.16 mm). The entire hippocampus is dissected from the remaining brain by a midline incision between the hemispheres and rolling the hippocampus out of the cerebral cortex. We have previously demonstrated the consistency of dissection of discrete brain regions in our laboratory (Bice et al., 2008; Kimpel et al., 2007; Liang et al., 2003). All equipment used to obtain tissue was treated with RNAse Zap (Ambion, Austin, TX) to prevent RNA degradation.
Isolation of RNA
Total RNA used for the qRT-PCR was isolated using the Trizol reagent combined with RNeasy mini kit (Qiagen, Valencia, CA). Concentration is determined by measuring the absorbance at A260 in a spectrophotometer. The purity was determined by the A260/A280 ratio. In addition, samples were run on agarose gels to confirm that the ribosomal bands were intact, indicating that the RNA was of high quality. To avoid DNA genomic contamination in the qRT-PCR assay, the RNA was treated with DNase I.
Microarray
The microarray study provided a means of prioritizing candidate genes within the 2 cM region. Each GeneChip® was scanned using an Affymetrix Model 3000 scanner and underwent image analysis using Affymetrix GCOS software. Raw .cel files were then imported into the statistical programming environment R (R Development Core Team, 2006) for further analysis with tools available from the Bioconductor Project. Quality assurance analysis was conducted using the BioConductor package affyQCReport (Gautier, et al., 2004; Miller, 2005; http://bioinformatics.picr.man.ac.uk/simpleaffy/index.jsp). Expression data were normalized and log2 transformed using the Robust Multi-chip Average (RMA) method (Bolstad et al., 2003; Irizarry et al., 2003) implemented in the Bioconductor package RMA. To increase power and decrease the false discovery rate, probe sets not reliably detected on at least one third of the microarrays in at least one experimental group in one of the three brain regions (using the Affymetrix Microarray Analysis Suite 5.0 detection call) were not analyzed (McClintick et al., 2006). Data from the three brain regions from each animal were averaged and the average value was analyzed to detect differences that were more general across multiple regions and thus more likely to reflect constitutive genetic differences as opposed to secondary effects. Linear modeling to calculate gene-wise p values was performed using the package Limma (Smyth, 2004). Mapping of probe sets to chromosomal locations was accomplished with data provided by Affymetrix. Calculated p values of probe sets mapping within the chromosome 10 candidate QTL region between 11,500,000 and 15,600,000 base pairs (locations of the microsatellite markers (D10Rat184 and D10Rat50) were then corrected for false discovery rate (FDR) by the method of Storey (Storey & Tibshirani, 2003). Probe sets were considered to be differentially expressed if the FDR adjusted p-value was p <= 0.10 (FDR 10%).
Quantitative real-time PCR
Because the linkage analyses and fine mapping studies were carried out in F2 animals that were derived from the non-inbred HAD1/LAD1 rats, we used animals from those selected lines to perform qRT-PCR. Using the ABI 7300 Real Time PCR System (Applied Biosystems, Foster City, CA), the relative mRNA expression levels of Crebbp, Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3 for the HAD1 and LAD1 rats were determined in the pre-frontal cortex, nucleus accumbens, amygdala, hippocampus, and caudate putamen by qRT-PCR. For the Crebbp gene, which was not differentially expressed in microarray, cDNA was generated from RNA for 6 HAD1 rats and 6 LAD1 rats (12 rats total) for each of the five brain regions. Also for this gene, we used an alternative qRT-PCR method in which cDNA was generated by pooling RNA from each individual brain region of 6 HAD1 rats and 6 LAD1 rats. We compared both qRT-PCR methods to determine if pooling would be sufficient to examine multiple brain regions and multiple genes. We found that both methods yielded very similar results. Therefore, we used the pooling method for the remaining genes (Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3), which had been differentially expressed in microarray and were confirmation studies. For all experiments, Superscript III First-Strand Synthesis System was used to perform the RT-PCR (Invitrogen, Carlsbad, CA). cDNA template, generated from 35 ng total RNA, was added to each PCR reaction that contained 0.20 μM forward and reverse primers and SYBR® Green PCR Master Mix (Invitrogen, Carlsbad, CA). For the individual samples, each experiment was performed in triplicate. For the pooled samples, each experiment was performed twice and each reaction was performed in triplicate. Vector NTI (Invitrogen) was used to design the forward and reverse primers for qRT-PCR (Table I). Each sample was amplified for 40 cycles, and the cycle threshold (Ct) was determined for each cDNA template. The Ct refers to the cycle number at which the fluorescence of the amplified product reached an arbitrary threshold that was within the exponential phase of amplification. To correct for sample-to-sample variation, an endogenous control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was amplified with the target and served as an internal reference to normalize the data. The average GAPDH Ct values for the HAD1 and LAD1 were the same in each brain region tested, making this an appropriate control gene to normalize the expression of the candidate genes of interest. Relative quantification of mRNA was performed using the standard curve method (Applied Biosystems, absolute quantitation guide; http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_041436.pdf). The ratio of a study gene (any 1 of the 6) to GAPDH is referred to as the normalization value. For each individual gene, the brain region with the lowest average normalization value was divided by itself (or set to 1.0), and it was divided by the average normalization values of each of the other brain regions to obtain a measure of relative expression for each region for that particular gene. t-Tests were performed to detect significant differences in expression between the HAD1 and LAD1 samples. Additionally, dissociation-curve analysis was examined to ascertain that non-specific amplification in the cDNA samples did not occur.
Table I.
List of Oligos
Quantitative Real-Time PCR
|
Results
Genetic Analyses of the HAD1 × LAD1 and HAD2 × LAD2 F2
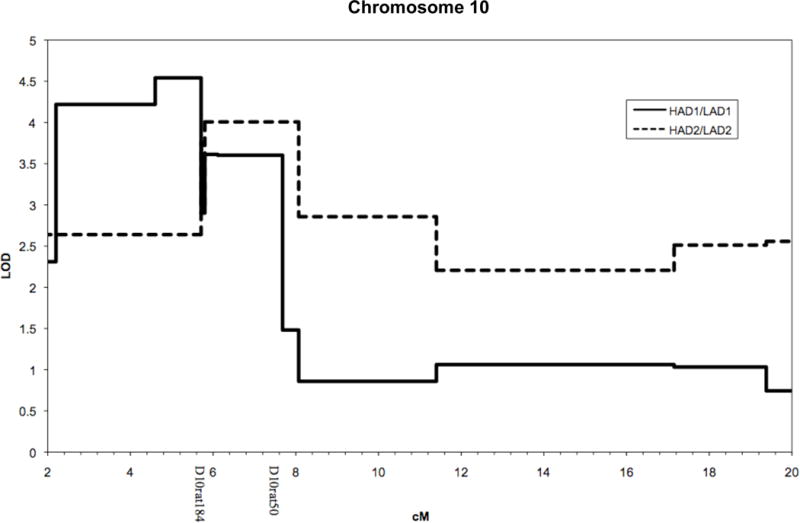
Initially, using 459 HAD1 × LAD1 F2 animals, Foroud and colleagues (2000) found evidence for linkage on chromosome 10 between the markers D10mgh25 and D10Mit16, a region spanning 18 cM. In a later study, new microsatellite markers were placed in and around the QTL region, providing additional support for the QTL. Analysis of additional markers as well as genotyping of the 8 inbred strain ancestors resulted in a LOD score of 4.54 on chromosome 10 near D10rat218 in the HAD1x LAD1 F2 sample. The data best fit an additive model and importantly narrowed the linkage peak to only 5.5 cM (2.2 – 7.7 cM; Figure I).
Fig. I.
Results of genetic analysis in the HAD1xLAD1 (solid line) and HAD2xLAD2 (dashed line) on chromosome 10. The Y-axis gives the LOD scores under an additive model. The X-axis gives the chromosomal position of the analyzed SNPs. Two markers (D10Rat184 and D10Rat50) on the X-axis show the location of the narrowed 2 cM region.
Genotyping of additional markers in the HAD2 × LAD2 F2 sample and the 8 inbred strain ancestors yielded a maximal LOD score of 4.01 with the additive model. The QTL interval was narrowed to 2.4 cM (5.8 – 8.1 cM), and the region overlapped with that identified by the analyses of the HAD1 × LAD1 F2 sample (Figure I). The overlapping region is 2 cM, 5.8 – 7.7 cM (11.5 to 15.6 Mb), and represents the critical interval examined in the present study.
mRNA expression in the HAD1 and LAD1 rats
qRT-PCR was used to determine relative mRNA expression levels for 6 genes within the 2 cM critical interval (11.5 to 15.6 Mb) on chromosome 10. Five of these genes ((Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3) demonstrated differential expression (FDR <= 0.10) in the average of three brain regions (nucleus accumbens, amygdala, and hippocampus) between the iHAD1 and iLAD1 using micro-array technology (see Table II).
Table II.
Microarray data for iHAD1 and iLAD1 rats. These genes are located in the narrowed 2 cM QTL region, from 11.5 to 15.6 Mb The significance value for the false discover rate (FDR) is p < 0.10.
| Airy Probeset | Gene Symbol | Gene Description | Chr. (q12) | Start | Fold Change | FDR p-value | Mean iHAD | Mean iLAD |
|---|---|---|---|---|---|---|---|---|
| 1374467_at | Trap1 | TNF receptor-associated protein 1 | 10 | 11,757,049 | −1.06 | 0.029 | 2092 | 2225 |
| 1386622_at | Gnptg | N-acetylglucosamine-1-phosphotransferase. γ subunit | 10 | 14,482,212 | 1.13 | 0.098 | 158 | 140 |
| 1369700_at | Clcn7 | chloride channel 7 | 10 | 14,379,802 | 1.10 | 0.015 | 455 | 414 |
| 1372676_at | Fahd1 | fumarylacetoacetate hydrolase domain containing 1 | 10 | 14,101,620 | 1.06 | 0.081 | 311 | 767 |
| 1398214_at | MapK8ip3 | mitogen-activated protein kinase 8 interacting protein 3 | 10 | 14,163,470 | 1.11 | 0.079 | 138 | 124 |
Fold change is the ratio of iHAD/iLAD.
Microarray results were derived using biological replicates; thus, significance for microarray results reflect mean differences and account for both biological and microarray technical variance. qRT-PCR results were derived from different, independent biological samples because microarray was performed on inbred samples and the authors wanted samples from selected lines to better corroborate with QTL data obtained from selected lines. In the case of Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3, we were confirming microarray results on different biological samples; therefore, for budgetary reasons, we elected to pool samples for qRT-PCR. Summary qRT-PCR statistics for these genes represent mean differences in expression with corresponding p-values accounting only for technical variance. Hence, we report technical confirmation of differential expression for these genes with a very low level of certainty. In addition, there is biological confirmation of differential expression in that we report replication of microarray results. In the case of Crebbp, however, which did not demonstrate statistically significant differential expression in the microarray experiment, qRT-PCR was obtained from multiple biological replicates and the reported significance level accounts for both biological and technical variance.
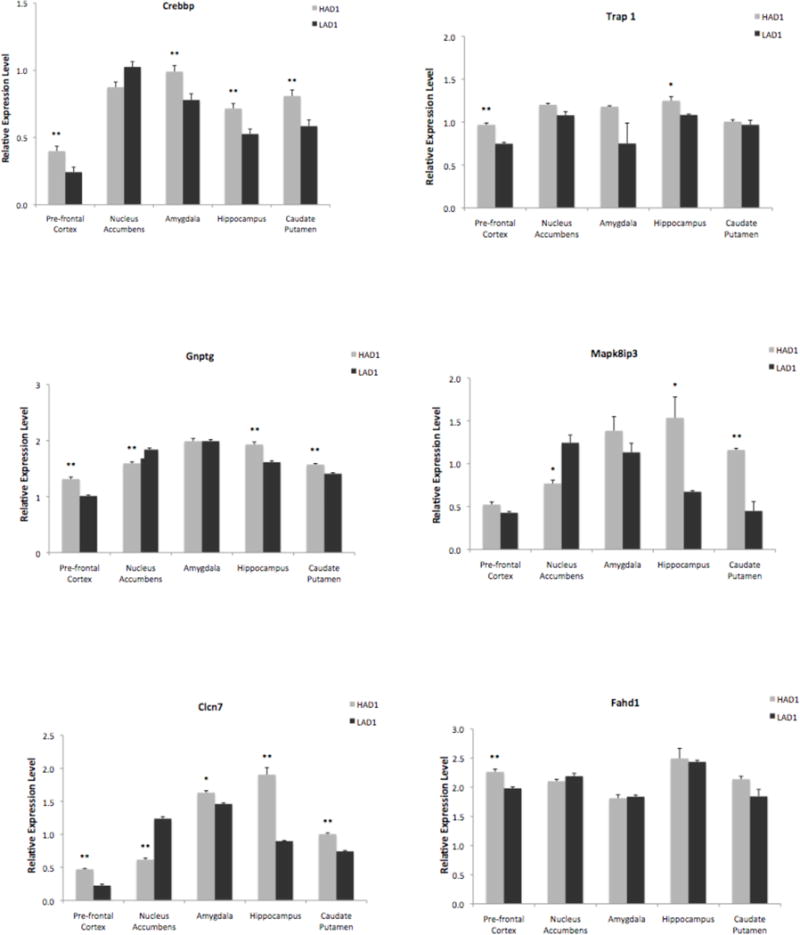
Relative mRNA expression levels can be seen in Figure II. The HAD1 rats showed significantly greater expression of the Trap1 gene in the pre-frontal cortex (p < 0.001) and hippocampus (p < 0.01) than the LAD1 rats. For Gnptg, the HAD1 rats showed significantly greater expression in the pre-frontal cortex (p < 0.001), hippocampus (p < 0.001), and caudate putamen (p < 0.001) but lower expression in the nucleus accumbens (p < 0.001) compared to the LAD1 rats. The Clcn7 gene was also differentially expressed in all brain regions tested. The HAD1 had significantly greater expression of Clcn7 than the LAD1 rats in the pre-frontal cortex (p < 0.001), amygdala (p < 0.01), hippocampus (p < 0.001), and caudate putamen (p < 0.001) but lower expression in the nucleus accumbens (p < 0.001). For the Fahd1 gene, the HAD1 rats showed significantly greater expression in the pre-frontal cortex (p < 0.001) than the iLAD1 rats. Expression of Mapk8ip3 was significantly greater for the HAD1 rats in the hippocampus (p < 0.01) and caudate putamen (p < 0.001) than the LAD1 rats but lower in the nucleus accumbens (p < 0.01). The Crebbp gene was differentially expressed in four of the five brain regions tested. The HAD1 had significantly greater expression of Crebbp in the pre-frontal cortex (p < 0.001), amygdala (p < 0.001), hippocampus (p < 0.001), and caudate putamen (p < 0.001) than the LAD1 rats. The fact that some genes show higher expression in some brain regions and lower expression in other brain regions is not unexpected. A gene’s function may vary depending on the brain region; the gene may play a more important role with regard to our phenotype in some brain regions than others.
Fig. II.
The figures show relative expression levels of six individual genes (Crebbp, Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3) in the HAD1 and LAD1 in five brain regions. The graph depicts the mean + S.E.M. of mRNA level. Y-axis represents the relative expression levels normalized to Gapdh. * Indicates a significant difference at p < 0.01; ** indicates a significant difference at p < 0.001 between the HAD1 and LAD1 as analyzed by Student’s t-test. For all genes, except for Crebbp, significance levels account for observed technical variance of repeated qRT-PCR measurements on pooled biological samples (see Results for a full explanation).
Discussion
In this study, we employed a powerful analytic approach that used ancestral recombination as well as F2 recombination to narrow a QTL on distal chromosome 10 in both the HAD1/LAD1 and HAD2/LAD2 lines. We identified a 2 cM region spanning from 11.5 to 15.6 Mb that overlaps in the two lines and represents our critical interval. To assess the molecular mechanisms underlying the QTL, we examined mRNA expression of six candidate genes within this critical interval. Based on the current alcohol literature, of the six genes examined herein, Crebbp and Mapk8ip3 may be the most promising with regard to alcoholism.
Using qRT-PCR, we found that the HAD1 rats displayed significantly greater levels of Crebbp mRNA expression than the LAD1 rats in the pre-frontal cortex, amygdala, hippocampus, and caudate putamen. The Crebbp gene is an interesting candidate because it provides instructions for making the cyclic adenosine 3′5′-monophosphate (cAMP) -response element binding protein (CREB). CREB is a transcription factor that binds to a DNA sequence, called cAMP response elements (CRE), and increases or decreases transcription and the expression of certain genes (Meyer & Habener, 1993). After the phosphorylation by protein kinase A (PKA) or Ca2+, CREB can modulate the expression of cAMP-inducible genes by binding to the promoter region of genes that contain CRE binding domains (Meyer and Habener, 1993; Pandey et al., 1999; Pandey et al., 2001). The CREB signaling cascade is known to regulate the transcription of tyrosine hydroxylase and many neuropeptides. The cAMP second messenger pathway has been shown to be important in mediating physiological and behavioral responses to ethanol (Pandey et al., 2001). Asher and colleagues (2002) have shown that ethanol induces cAMP-dependent gene expression regulated by CREB and PKA, and they believe that the signaling pathway may mediate some of the addictive behaviors underlying alcoholism. Pandey et al. (1999) found that the CRE-DNA binding activity in the amygdala of the alcohol preferring (P) rats may be associated with the higher level of anxiety observed in these animals. Also, it has been suggested that CREB may be important in the formation of long-term memories (Alarcon et al., 2004; Cooke and Bliss, 2005; Korzus et al., 2004; Martin and Sun, 2004; Wood et al., 2005), which may ultimately be vital for understanding addiction. As Nestler (2001) has pointed out, addiction may be viewed as a form of drug-induced neural plasticity. Because drugs of abuse have been shown to produce morphological changes in particular neuronal cell types after chronic drug administration, these structural changes may be a type of consolidation, a form of neuronal adaptation analogous to learning and memory, which may be required for the long-term effects associated with the addictive process (Nestler, 2001).
Mapk8ip3, mitogen-activated protein kinase 8 interacting protein 3, is another potentially important candidate gene located within the critical interval on chromosome 10. The HAD1 rats displayed significantly higher levels of Mapk8ip3 mRNA expression in the hippocampus and caudate putamen than the LAD1 rats but significantly lower levels in the nucleus accumbens. MAP kinases (MAPks) are evolutionarily conserved regulators that mediate signal transduction and play essential roles in various physiological processes (Zhang and Dong, 2007). Recent evidence suggests a role for the MAPK family in several effects of ethanol. In an extensive review, Aroor and Shukla (2004) present evidence that ethanol influences MAPK in diverse cell/organ systems and exhibit different patho-physiological consequences, such as liver injury, pancreatitis, and neuronal toxicity. Interestingly, MAPK has an important association with the cAMP signaling cascade. Constantinescu et al (2004) found that the inhibition of the MAPK pathway enhances cAMP-dependent gene activation during exposure of cells to ethanol. MAPK signaling appears to increase CRE-mediated gene expression by removing the restraint of CREB binding protein (CBP) on CREB transcriptional activity (Constantinescu et al., 2004). The MAPK family could also play an important role in the alcohol induced changes in synaptic plasticity associated with the effects of alcohol abuse on learning and memory (Roberto et al., 2003). It has been shown that CREB activation via the MAPK cascade may influence long-term memory formation (Cooke and Bliss, 2005). Further, the protein encoded by Mapk8ip3 may interact with and regulate the activity of numerous protein kinases of the c-Jun NH2-terminal kinase (JNK) signaling pathway, and thus function as a scaffolding protein in neuronal cells and as an adaptor protein for axonal vesicular transport (Kelkar et al., 2003). The JNK signaling pathway is also activated in response to many stressful stimuli, including ethanol-induced cell death (Han et al., 2008; Ku et al., 2007).
Although it is currently unclear as to what role, if any, the Trap1, Gnptg, Clcn7, and Fahd1 genes play with regard to alcohol intake, each appear to underlie complex functions, and their influence may not be obvious at this time. For example, the Trap1 gene is a mitochondrial HSP90 protein that is a highly conserved molecular chaperone that plays a key role in signal transduction, protein folding, protein degradation, and morphologic evolution (Pagliarini et al. 2008). HSP90 proteins normally associate with other chaperones and are important for folding newly synthesized proteins or stabilizing and refolding denatured proteins after stress (Wegele et al. 2004).
The Clcn7 gene showed significant differences in expression between the HAD1 ad LAD1 rats in all brain regions tested. This gene encodes chloride channel 7, which produces proteins that belong to the CLC chloride channel family. Chloride channels play important roles in the plasma membrane and in intracellular organelles. Defects in Clcn7 have been shown to cause osteopetrosis in both men and mice (Kornak et al. 2001). Using the alcohol preferring (P) and non-preferring (NP) rats and the HAD1/LAD1 and HAD2/LAD2 rats, Alam et al. (2005) found that there is no consistent relationship between preference for alcohol consumption and high bone mass or strength. However, it appears that genes that regulate bone mass and strength segregate with alcohol preference genes in the P and HAD rat lines, suggesting that alcohol preferring rat lines may be useful for identifying genes that regulate bone mass and structure.
The Gnptg gene provides instructions for making one the gamma subunits of an enzyme called GlcNAc-1-phosphotransferase, which helps prepare certain newly made enzymes for transport to lysosomes (Braulke et al. 2008), which are compartments within the cell that use digestive enzymes called hydrolases to break down large molecules into smaller ones that can be reused by the cell. The Fahd1 gene encodes fumarylacetoacetate hydrolase (FAH), an enzyme abundant in the liver and kidneys. FAH is the last in a series of five enzymes needed to break down the amino acid tyrosine, a protein building block found in many foods. Specifically, FAH converts a tyrosine byproduct called fumarylacetoacetate into smaller molecules that are either excreted by the kidneys or used in reactions that produce energy.
In the present study, we identified a QTL region on distal chromosome 10 that was in common in the HAD1/LAD1 and HAD2/LAD2 analyses. We examined mRNA expression of six genes within this region. Of the six genes tested, we believe Crebbp and Mapk8ip3 may be promising candidates with regard to alcohol drinking. Both genes show differential expression between the HAD1 and LAD1 rats in brain regions that are believed to be important for alcohol-related behavior and have been recognized as targets for the effects of ethanol exposure in the brain and/or other organ systems (Aroor and Shukla, 2004; Pandey et al., 2001). Future studies will examine each of the six genes to determine if there is sequence variation between the HAD and LAD rats.
Acknowledgments
This work was supported by a grant from the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health (Grant R01 AA015933 and R01AA10707). We wish to thank William J. McBride and Mark Kimpel for sharing the microarray data from the iHAD1/iLAD1 rat study. We especially thank Stephany Bice for her work in assembling the reference section.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam I, Robling AG, Weissing S, Carr LG, Lumeng L, Turner CH. Bone mass and strength: phenotypic and genetic relationship to alcohol preference in P/NP and HAD/LAD rats. Alcohol Clin Exp Res. 2005;29:1769–76. doi: 10.1097/01.alc.0000183005.28502.4f. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Aroor AR, Shukla SD. MAP kinase signaling in diverse effects of ethanol. Life Sci. 2004;74:2339–2364. doi: 10.1016/j.lfs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Asher O, Cunningham TD, Yao L, Gordon AS, Diamond I. Ethanol stimulates cAMP-responsive element (CRE)-mediated transcription via CRE-binding protein and cAMP-dependent protein kinase. J Pharmacol Exp Ther. 2002;301:66–70. doi: 10.1124/jpet.301.1.66. [DOI] [PubMed] [Google Scholar]
- Bice PJ, Liang T, Zhang L, Strother WN, Carr LG. Drd2 expression in the high alcohol-preferring and low alcohol-preferring mice. Mamm Genome. 2008;19:69–76. doi: 10.1007/s00335-007-9089-2. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high-density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Braulke T, Pohl S, Storch S. Molecular analysis of the GlcNac-1-phosphotransferase. J Inherit Metab Dis. 2008;31:253–257. doi: 10.1007/s10545-008-0862-5. [DOI] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Carr LG, Habegger K, Spence J, Ritchotte A, Liu L, Lumeng L, Li TK, Foroud T. Analyses of quantitative trait loci contributing to alcohol preference in HAD1/LAD1 and HAD2/LAD2 rats. Alcohol Clin Exp Res. 2003;27:1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- Constantinescu A, Wu M, Asher O, Diamond I. cAMP-dependent protein kinase type I regulates ethanol-induced cAMP response element-mediated gene expression via activation of CREB-binding protein and inhibition of MAPK. J Biol Chem. 2004;279:43321–43329. doi: 10.1074/jbc.M406994200. [DOI] [PubMed] [Google Scholar]
- Cooke SF, Bliss TV. Long-term potentiation and cognitive drug discovery. Curr Opin Investig Drugs. 2005;6:25–34. [PubMed] [Google Scholar]
- Foroud T, Bice P, Castelluccio P, Bo R, Miller L, Ritchotte A, Lumeng L, Li TK, Carr LG. Identification of quantitative trait loci influencing alcohol consumption in the high alcohol drinking and low alcohol drinking rat lines. Behav Genet. 2000;30:131–140. doi: 10.1023/a:1001955205117. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Han JY, Jeong EY, Kim YS, Roh GS, Kim HJ, Kang SS, Cho GJ, Choi WS. C-jun N-terminal kinase regulates the interaction between 14-3-3 and Bad in ethanol-induced cell death. J Neurosci Res. 2008;86:3221–3229. doi: 10.1002/jnr.21759. [DOI] [PubMed] [Google Scholar]
- Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res. 1984;8:477–479. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high-density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kelkar N, Delmotte MH, Weston CR, Barrett T, Sheppard BJ, Flavell RA, Davis RJ. Morphogenesis of the telencephalic commissure requires scaffold protein JNK-interacting protein 3 (JIP3) Proc Natl Acad Sci USA. 2003;100:9843–9848. doi: 10.1073/pnas.1733944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornak U, Kasper D, Bösl MR. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 2001;104:205–15. doi: 10.1016/s0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku BM, Lee YK, Jeong JY, Mun J, Han JY, Roh GS, Kim HJ, Cho GJ, Choi WS, Yi GS, Kang SS. Ethanol-induced oxidative stress is mediated by p38 MAPK pathway in mouse hippocampal cells. Neurosci Lett. 2007;419:64–67. doi: 10.1016/j.neulet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L. Alcohol preference and voluntary alcohol intakes of inbred rat strains and the National Institutes of Health heterogeneous stock of rats. Alcohol Clin Exp Res. 1984;8:485–486. doi: 10.1111/j.1530-0277.1984.tb05708.x. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23:163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, Lumeng L, Li TK, Foroud T, Carr LG. alpha-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci USA. 2003;100:4690–4695. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Sun YE. To learn better, keep the HAT on. Neuron. 2004;42:879–881. doi: 10.1016/j.neuron.2004.06.007. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by present call on analysis of microarray experiments. BMC Bioinformat. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer TE, Habener JF. Cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr Rev. 1993;14:269–290. doi: 10.1210/edrv-14-3-269. [DOI] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Molecular neurobiology of addiction. Am J Addict. 2001;10:201–217. doi: 10.1080/105504901750532094. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Mittal N, Lumeng L, Li TK. Involvement of the cyclic AMP-responsive element binding protein gene transcription factor in genetic preference for alcohol drinking behavior. Alcohol Clin Exp Res. 1999;23:1425–1434. [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Saito T, Yoshimura M, Sohma H, Götz ME. cAmp signaling cascade: a promising role in ethanol tolerance and dependence. Alcohol Clin Exp Res. 2001;25:46S–48S. doi: 10.1097/00000374-200105051-00008. [DOI] [PubMed] [Google Scholar]
- Paxinos GW. The rat brain in stereotoxic coordinates. 4th. San Diego, CA: Academic Press; 1998. [Google Scholar]
- Roberto M, Nelson TE, Ur CL, Brunelli M, Sanna PP, Gruol DL. The transient depression of hippocampal CA1 LTP induced by chronic intermittent ethanol exposure is associated with an inhibition of the MAP kinase pathway. Eur J Neurosci. 2003;17:1646–1654. doi: 10.1046/j.1460-9568.2003.02614.x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Spuhler K, Deitrich RA. Correlative analysis of ethanol-related phenotypes in rat inbred strains. Alcohol Clin Exp Res. 1984;8:480–484. doi: 10.1111/j.1530-0277.1984.tb05707.x. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Nat Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Louis M, Tanguay RM. Mutations in the fumarylacetoacetate hydrolase gene causing hereditary tyrosinemia type I: overview. Hum Mutat. 1997;9:291–9. doi: 10.1002/(SICI)1098-1004(1997)9:4<291::AID-HUMU1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Wegele H, Müller L, Buchner J. Hsp70 and Hsp90–a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dong C. Regulatory mechanisms of mitogen-activated kinase signaling. Cell Mol Life Sci. 2007;64:2771–2789. doi: 10.1007/s00018-007-7012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


