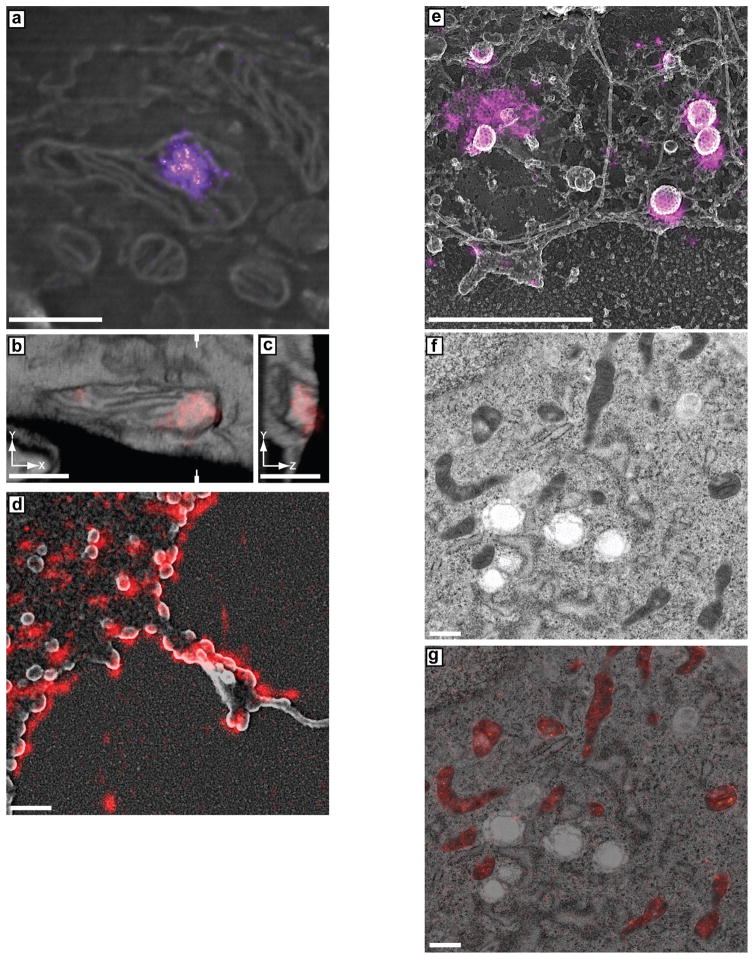
Figure 6.
Examples of anticipated results. (a–c) Examples of CLEM using Tokuyasu cryosectioning adapted with permission from Kopek et al.18. NIH3T3 mouse embryonic fibroblast cells were transfected with a plasmid expressing the mitochondrial DNA-binding protein (mitochondrial transcription factor A (TFAM)) genetically fused to the paFP mEos2, rendered in red or magenta. (a) Example result for 2D CLEM imaging of mitochondrial nucleoids using the Tokuyasu protocol. (b,c) Example result for 3D CLEM of mitochondrial nucleoids using the Tokuyasu protocol with iPALM and FIB-SEM showing (b) an x–y plane and (c) a y–z plane sectioned through the volume as indicated by the marks in b. (d) Example results for whole-cell-mount method showing correlated iPALM and SEM of COS7 cells transfected with a plasmid expressing the HIV Gag protein fused to mEos2. (e) Platinum replica of an unroofed HeLa cell correlated with dSTORM of the Alexa-Fluor-647-labeled clathrin adaptor, AP-2. (f,g) Example of transmission electron micrograph (f) overlaid with a PALM (red/white) image (g) of NIH3T3 mouse embryonic fibroblast cells transfected with a plasmid expressing a mitochondrial matrix localized mEos4a. The sample was high-pressure frozen, freeze-substituted with 0.5% OsO4, and embedded in GMA resin. Scale bars, 500 nm.