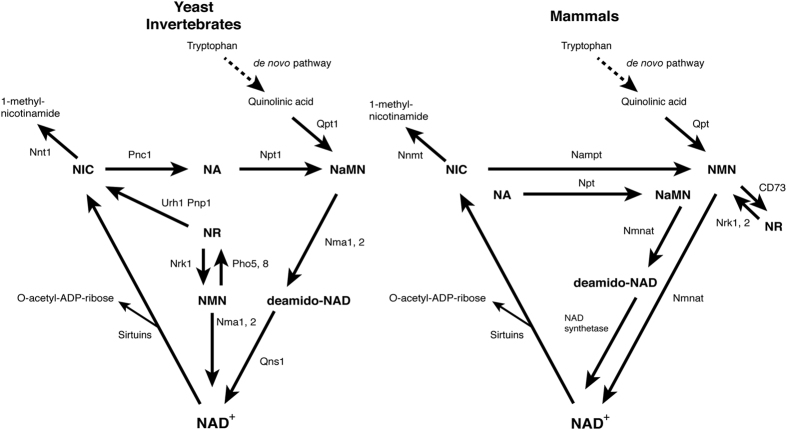
Figure 1.
Comparison of NAD+ biosynthetic pathways between yeast/invertebrates and mammals. Left panel: NAD+ biosynthetic pathways in the budding yeast Saccharomyces cerevisiae and invertebrates. The pathways from NIC, NA, and NR and the de novo pathway from tryptophan are shown. Right panel: NAD+ biosynthetic pathways in mammals. In mammals, NIC is a predominant NAD+ precursor. NA, NR, and tryptophan can also be utilized to synthesize NAD+. The de novo pathway and the NAD+ biosynthetic pathway from NA are evolutionarily conserved, whereas the NAD+ biosynthetic pathway from NIC is mediated by NAMPT. In this figure, only sirtuins are shown among multiple NAD+-consuming enzymes that break NAD+ into nicotinamide and ADP-ribose. Mammals have two NR kinases (Nrk1 and 2) and ecto-5′-nucleotidase CD73 to produce NMN and NR, respectively. NA, nicotinic acid; NAD+, nicotinamide adenine dinucleotide; NAMPT, nicotinamide phosphoribosyltransferase; NIC, nicotinamide; NR, nicotinamide riboside; Nrk1, nicotinamide ribose kinase 1; Npt1, nicotinic acid phosphoribosyltransferase; Nma1, 2, nicotinic acid mononucleotide adenylyltransferase 1, 2; Nnt1, nicotinamide-N-methyltransferase; Npt, nicotinic acid phosphoribosyltransferase; NMNAT, NMN adenylyltransferase; NNMT, nicotinamide-N-methyltransferase; NaMN, nicotinic acid mononucleotide; NMN, nicotinamide mononucleotide; Pho5, 8, phosphatase 5, 8; Pnp1, purine nucleoside phosphorylase; Pnc1, nicotinamidase; Qpt, quinolinic acid phosphoribosyltransferase; Qns1, NAD synthetase; Qpt1, quinolinic acid phosphoribosyltransfease; Urh1, uridine hydrolase.