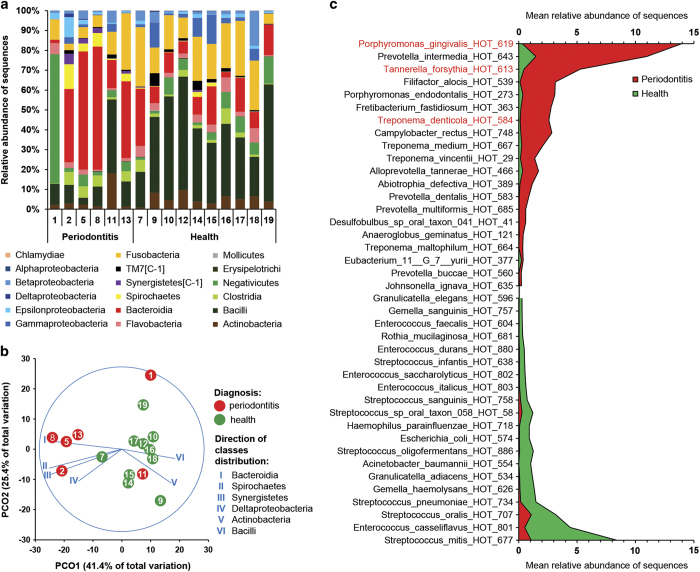
Figure 1.
Composition of microbial communities from periodontal pockets as assessed by metatranscriptome analysis. (a) Composition of the periodontal community in the 16 individuals shown on the class level. Healthy subjects and those diagnosed with periodontitis are grouped on the right and left side of the subpanel, respectively. (b) Principal coordinates analysis (PCoA) of periodontal communities. Bray–Curtis similarity values were calculated on standardised log2 transformed abundances of reads grouped to Classes. Communities from healthy individuals and those with periodontitis are marked in green and red, respectively. Vectors represent the direction of the relationships between classes and ordination axes (see Materials and Methods for details). (c) Transcriptionally active species associated with health and disease. Relative abundance in health and disease of reads grouped to species (human oral taxons, HOTs) that were associated with periodontitis (upper part) and health (lower part). ‘Red-complex’ species are highlighted in red. P<0.05 for all data shown.