Figure 1.
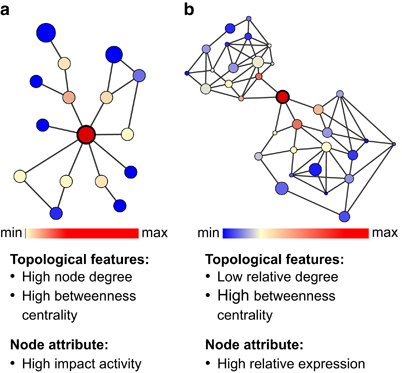
Criteria for defining keystone nodes in microbial species interaction and community-wide metabolic networks. (a) Criteria for identifying keystone species in reconstructed species interaction networks. Nodes represent taxa and edges represent associations between them. Node sizes reflect activity. (b) Criteria for identifying genes encoding key functionalities in reconstructed community-wide metabolic networks. Nodes represent enzyme-coding genes and edges correspond to shared metabolites (either reactants, products or educts). Node sizes reflect relative expression.
