Figure 1. TAP-tagged RNA exosome complex in antigen activated B cells is localized in the nucleus and associates with RNA/DNA helicase activity of Mtr4.
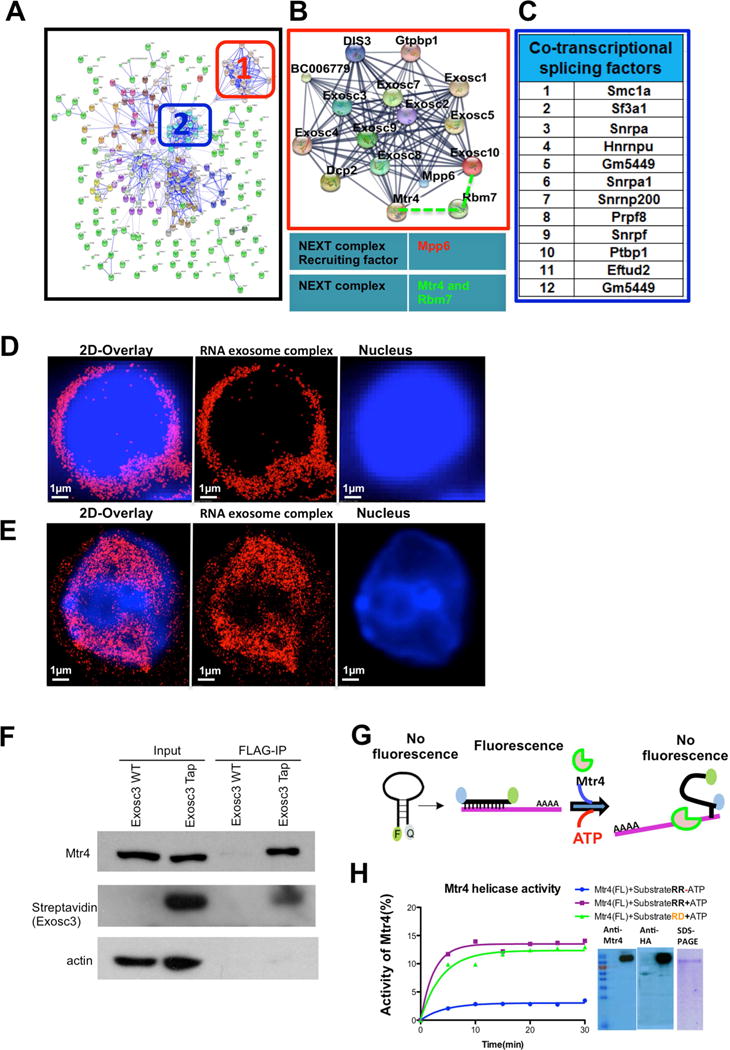
(A) Interaction network of RNA exosome complex associated proteins. In-silico network analysis of proteins identified by mass-spectrometry was performed using the STRING database combined with experimental data. (B) Higher magnification view of the boxed region (1) shows RNA exosome complex associating with NEXT complex components Mtr4 and RBM7 along with Mpp6 (recruiter of the NEXT complex), respectively. (C) Identification of the most stringent interacting partners shown in panel A cluster 2; these proteins are related with cotranscriptional splicing pathways. (D) Reconstructed single color 3D-STORM image from a data set of 50,000 frames of fixed splenic B cells before CSR stimulation, in which Exosc3 was labeled with AlexaFluor647 and the nucleus with DAPI. (E) Single color 3D STORM image of B cells after CSR stimulation, in which Exosc3 was labeled with AlexaFluor647 and the nucleus with DAPI. (F) Flag immunoprecipitation reactions (IPs) were performed on Exosc3TAP B cells to demonstrate Exosc3 and Mtr4 interaction. (G) Schematic representation of molecular beacon (6-FAM and Iowa Black) based helicase assay for Mtr4 protein. (H) Two substrates (RNA-RNA and RNA-DNA) were designed with 5′–6FAM and 3′Iowa black linked to 17mer RNA molecule, which annealed with 22mer RNA and DNA complimentary strand to evaluate the helicase activity of Mtr4 for both substrates. hMtr4 full length protein was expressed in HEKA-293T cells and purified by affinity tag. Expression of the protein was confirmed by western blot using anti Mtr4 antibodies. Percentage activity of hMtr4(FL) was calculated by pseudo-first order rate constant describing fluorescence decay upon ATP addition [[k (min-1)]. All reactions were performed in triplicate, and error bars represent standard deviation among independent reactions. All of the 3D STORM imaging was performed in three different B cells (from independent experiments) and repeated three or more times. 3D STORM super resolution image magnification is x100. Scale bars: 1μm(D) &(E). Also, see supplementary video 1 and 2.
