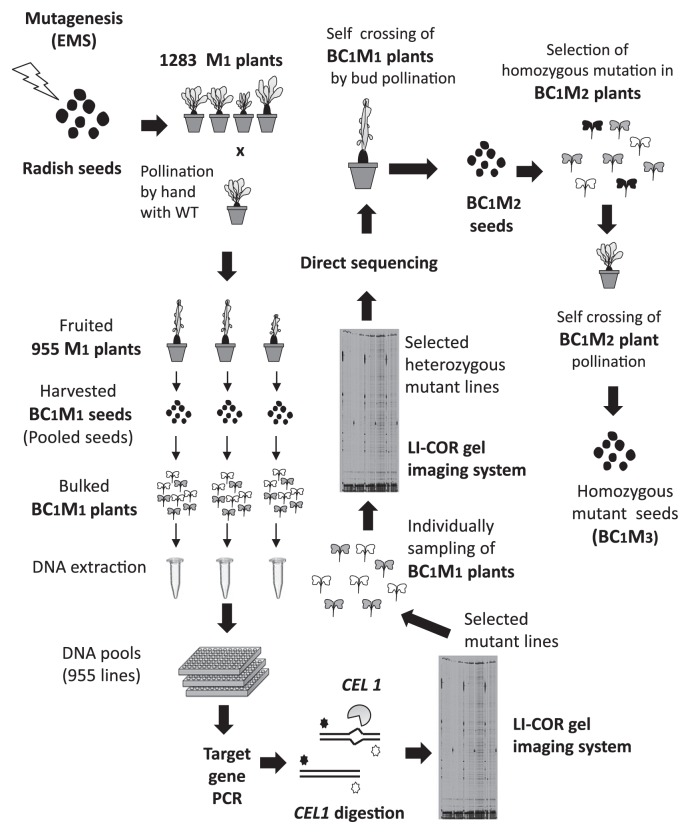
Fig. 3.
The Radish-TILLING platform. In this TILLING system, radish seeds were mutagenized with the chemical mutagen EMS to produce M1 plants. The M1 plants were crossed with wild-type plants because radish is self-incompatible, producing 955 BC1M1 lines. DNA was extracted from eight bulked BC1M1 plants per line for analysis. The target DNA was amplified using a forward primer labeled with 700 nm dye and a reverse primer labeled with 800 nm dye. The amplified PCR products were digested with CEL 1, and the resulting DNA fragments were separated and detected in the 700 and 800 dye channels of a LI-COR DNA Analyzer. Since the BC1M1 population (in bulk) was predicted to exhibit a mutation ratio of 1:3, a peak sequence signal corresponding to a mutation would have one-fourth the signal intensity of wild type. Therefore, it would be difficult to detect the mutated nucleotide by sequencing DNA from the BC1M1 population. To facilitate mutant identification, an individual BC1M1 plant was grown and analyzed again using TILLING gel analysis. A peak sequence signal corresponding to a mutation in the selected BC1M1 individual should have the same intensity as that of wild type. Selected BC1M1 individuals with heterozygous mutations were self-crossed by bud pollination. The homozygous mutant was selected based on the presence of a single peak in the sequencing signal representing a mutation. The population was then harvested as BC1M2 seeds from BC1M2 plants subjected to bud pollination.