Abstract
Voltage-sensing phosphatase (VSP) consists of a transmembrane voltage sensor and a cytoplasmic enzyme region. The enzyme region contains the phosphatase and C2 domains, is structurally similar to the tumor suppressor phosphatase PTEN, and catalyzes the dephosphorylation of phosphoinositides. The transmembrane voltage sensor is connected to the phosphatase through a short linker region, and phosphatase activity is induced upon membrane depolarization. Although the detailed molecular characteristics of the voltage sensor domain and the enzyme region have been revealed, little is known how these two regions are coupled. In addition, it is important to know whether mechanism for coupling between the voltage sensor domain and downstream effector function is shared among other voltage sensor domain-containing proteins. Recent studies in which specific amino acid sites were genetically labeled using a fluorescent unnatural amino acid have enabled detection of the local structural changes in the cytoplasmic region of Ciona intestinalis VSP that occur with a change in membrane potential. The results of those studies provide novel insight into how the enzyme activity of the cytoplasmic region of VSP is regulated by the voltage sensor domain.
Keywords: membrane potential, phosphoinositide, ion channel
Most proteins contain multiple functional domains, and domain-domain coupling is critical to proteins’ functionality. The voltage-sensor domain (VSD) is one of the best-characterized functional domains of membrane proteins. It has the crucial function of sensing transmembrane potential to regulate ion permeation through voltage-gated ion channels [1] and also functions in voltage-sensing phosphatases. Operation of the VSD is critical for many important physiological systems, including the nervous, cardiovascular and endocrine systems.
Within voltage-gated ion channels, the VSD is coupled to the pore domain or pore-gate domain (PGD). The PGD contains two transmembrane segments with a turret segment that provides an ion-selectivity filter governing ion permeation (Fig. 1a). The VSD consists of four transmembrane helical segments, of which the fourth segment, S4, is key for sensing membrane potential. Its signature sequence contains positively charged residues situated periodically and separated by pairs of intervening hydrophobic residues. Other helices comprising the VSD contain negatively charged residues that counterbalance with positive charges in S4. By changing the salt bridge partners (negatively charged residue) in the S1 to S3 helices, changes in membrane potential induce movement in the positively charged residues of S4. This results in translocation of the entire S4 segment relative to the other parts of the VSD. This movement of the VSD causes a structural change in the PGD, either through mechanical force via a linker segment or through helix-helix interaction within the membrane to effect opening or closing of the channel pore.
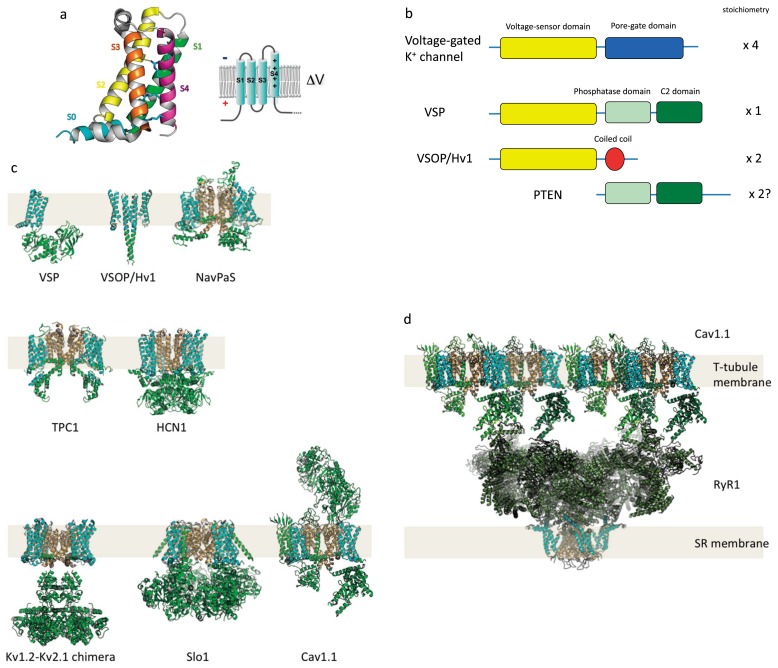
Figure 1.
Structure of VSD and domain organization of VSD-containing proteins.
(a) Structure of Ci-VSD, the voltage-sensor domain of Ci-VSP (Ciona intestinalis voltage-sensing phosphatase) (PDB ID: 4G7V). Right shows a membrane topology of VSD. S4 has multiple positive charges (+) critical for sensing transmembrane voltage. (b) scheme of domain organization of VSP as compared with other related proteins, including voltage-gated potassium channel, voltage-gated proton channel (VSOP/Hv1) and PTEN. (c, d) Atomic structures of VSD-containing proteins are shown as a gallery. VSP: Ci-VSP (full length model based on coordinates from PDB ID:3AWF and PDB ID: 4G7V), VSOP/Hv1: mouse voltage-gated proton channel (dimer model based on a coordinate from PDB ID: 3WKV), NavPaS: insect voltage-gated sodium channel (PDB ID: 5X0M), TPC1: plant two-pore cation channel (PDB ID: 5E1J), HCN1: human Hyperpolarization-activated cyclic nucleotide-gated channel (PDB ID: 5U6O), Kv1.2-Kv2.1 chimera: human voltage-gated potassium channel (PDB ID: 2R9R), Slo1: large conductance calcium activated voltage-gated potassium channel from Aplysia (PDB ID: 5TJ6), Cav1.1: human skeletal muscle-type voltage-gated calcium channel (PDB ID: 5GJV). VSP has a single VSD. VSOP/Hv1 has two VSDs in a dimer. NavAb, Kv1.2-Kv2.1 chimera and Slo1 have four VSDs in a tetramer. TPC1 has four VSDs in a dimer. Cav1.1 has four VSDs in a monomer. (d) Functional unit of excitation-contraction coupling in skeletal muscle cells, which consists of four Cav1.1 subunits and a tetrameric ryanodine receptor calcium-release channel, RyR1 (PDB ID: 3J8H). Auxiliary subunits of Cav1.1 are not shown here. In Cav1.1, motion of the VSD is translated into a structural change in the cytoplasmic domain to induce opening of the RyR pore. This interaction does not require calcium permeation of the Cav1.1 PGD, and direct interaction with the large cytoplasmic domain of RyR plays a central role in this electrochemical coupling. In all pictures, the VSD and PGD are shown as cyan and light orange, respectively.
In most voltage-gated ion channels, including the Shaker-type voltage-gated potassium (Kv) channel [2], bacterial voltage-gated sodium (Nav) channel [3] and L-type voltage-gated calcium (Cav) channel [4], the VSD is domain-swapped within tetramers, where the VSD is located close to the PGD of the neighboring subunit. In the high conductance, Ca2+ activated K+ channel (BK channel), [5], Eag K+ channel [6] and HCN1 hyperpolarization-activated cation channel [7], by contrast, the VSD is not domain-swapped and flanks the PGD in the same subunit. In addition, by interacting with other proteins, some voltage-gated ion channels can transmit signals deeper within cells. For instance, Cav channels directly interact with another type of calcium channel, the ryanodine receptor, on the sarcoplasmic reticulum (Fig. 1d) to activate calcium release from internal calcium stores. This enables rapid and simultaneous neural control of contraction of large vertebrate skeletal muscle cells.
Unlike conventional voltage-gated ion channels with tetrameric organizations or four homologous repeats in a single subunit, some VSD-containing proteins do not have a PGD. One such example is the voltage-gated proton channel VSOP/Hv1, within which the VSD plays dual voltage sensing and ion permeation roles [8–10]. VSOP/Hv1 forms a dimer through interaction between C-terminal coiled coils [11] and through interfaces of transmembrane helices [12,13] (Fig. 1c). Another VSD-containing protein that lacks a PGD is the voltage-sensing phosphatase (VSP) [14]. VSP contains a single VSD, which is connected to a cytoplasmic region that is remarkably similar to the tumor suppressor enzyme PTEN, which catalyzes dephosphorylation of phosphoinositides (Fig. 1c). Within VSP, membrane depolarization induces a structural change in the VSD that triggers activation of the cytoplasmic phosphoinositide phosphatase. VSP gene is conserved from marine invertebrates to humans. In the time since the first characterization of the sea squirt VSP (Ciona intestinalis VSP; Ci-VSP) [14], VSP orthologs have been characterized from a teleost [15], amphibian [16], chick [17] and mammals [18,19]. Human and rodent VSPs have also been characterized as PTEN-related phosphatases, called TPTE and TPIP [20–22].
As a VSD-containing protein, VSP has two unusual features. First, the effector region is the cytoplasmic enzyme, not a transmembrane domain such as the PGD of voltage-gated ion channels. So far VSP is the only established case where a cytoplasmic region with enzyme activity is regulated by a VSD. Second, VSP has a single VSD (Fig. 1c, 2a), in contrast with voltage-gated ion channels, where multiple VSDs regulate the downstream effector in a complex manner. Thus, VSP is a unique protein target with which to unravel the basic operating principles of the VSD and downstream effector and of the domain-domain coupling underlying signal transduction by proteins.
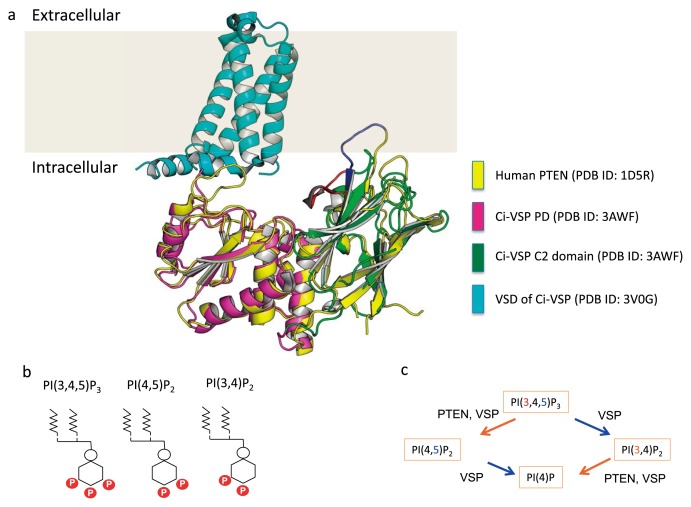
Figure 2.
VSP structure and enzyme activity
(a) Structure of the VSD and PTEN-like cytoplasmic region of VSP. The VSD of Ci-VSP (3V0G) is in cyan. The X-ray crystal structure of the cytoplasmic region of Ci-VSP (3AWF; green for C2 domain and magenta for PD (phosphatase domain)) is superimposed on the human PTEN structure (1D5R; yellow). Blue and red indicate distinct structure in C2 domain between PTEN and Ci-VSP. A loop of C2 domain of PTEN(blue) orients the membrane, whereas a loop of C2 domain of Ci-VSP (red) orients the phosphatase domain. Y522 that interacts with H332 in active center (See the text) is located in this loop. (b) VSP substrates include PtdIns(3,4,5)P3, PtdIns(3,4)P2 and PtdIns(4,5)P2. (c) VSP cleaves the 3-phosphate from PtdIns(3,4,5)P3 and PtdIns(3,4)P2 and 5-phosphate from PtdIns(3,4,5)P3 and PtdIns(4,5)P2. By contrast, PTEN removes the 3-phosphate from PtdIns(3,4,5)P3 and PtdIns(3,4)P2.
Structure of VSP
The C-terminal cytoplasmic region of VSP shares structural organization with PTEN (Fig. 2a). Both contain a phosphatase domain (PD) consisting mainly of α helices and loops, and a C2 domain consisting mainly of β-sheets and loops. The N-terminal segment of PTEN, which flanks the PD and is called the phosphoinositide binding motif (PBM), is also conserved in VSP and constitutes part of the linker between the VSD and PD (VSD-PD linker). However, VSP lacks a region corresponding to the C-terminal regulatory domain, which in PTEN is known to be phosphorylated and to regulate enzyme activity through direct association with the PD and C2 domain [23]. The N-terminus of VSP has a cytoplasmic stretch that varies in length and amino acid sequence among the ortholog proteins.
The X-ray crystal structures of both the VSD and C-terminal cytoplasmic enzyme region of Ci-VSP were solved as separate molecules (Fig. 1c, 2a). The overall structure of the VSP cytoplasmic region is superimposable on that of PTEN [24] (Fig. 2a). Despite this structural similarity, the substrate specificity of VSP differs from that of PTEN. VSP cleaves the 5-phosphate from phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3) and phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) [25,26] and the 3-phosphate of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 [27]. By contrast, PTEN exclusively cleaves the 3-phosphate from PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (Fig. 2b, c). The difference in substrate specificity between VSP and PTEN does not reflect the presence of the VSD, since a chimeric protein (VSPTEN) consisting of the VSD of VSP and PTEN [28], shows the same 3 phosphate specificity as PTEN. Details of the structural basis for the difference in substrate specificity between VSP and PTEN remain to be established.
The atomic structure of the VSD of Ci-VSP (Ci-VSD) was obtained using X-ray crystallography in combination with EPR analysis [29]. The molecular architecture of Ci-VSD is similar to the VSDs of voltage-gated ion channels. Multiple (four in the case of Ci-VSD) arginines form salt bridges with negatively charged residues in other helices from VSDs. Two aqueous cavities, each for extracellular side and intracellular side, are separated by a hydrophobic residue at the inner core and a charged residue in S4 must pass this shield to carry charge when the VSD is activated upon membrane depolarization. This scenario is supported by a study of state-dependent accessibility after histidine substitution of individual arginines in S4 [30]. In addition, at the N-terminal side of S1 is a short cytoplasmic helix, which runs parallel to the membrane, as is the case in many voltage-gated ion channels.
Several features of the structure of Ci-VSD differ from conventional voltage-gated ion channels. Within voltage-gated ion channels, a segment of the S4 helix in the core region of the VSD forms a salt bridge with other helices that form a 310 helix. By contrast, the corresponding region of Ci-VSD forms α-helix, not 310 helix. In addition, the negative charge on S2 within Ci-VSP contributes no countercharge for S4 and does not form a salt bridge with that segment, as is also the case in VSOP/Hv1. Lastly, fluorescent tagging revealed that the N-terminal cytoplasmic region of Ci-VSP moves upon a change in membrane potential [31], whereas mobility of the N-terminus of the VSD has not been reported in conventional voltage-gated ion channels.
Wild-type Ci-VSP is not fully activated at around 0 mV [14], but the voltage-dependence of a Ci-VSP with mutation of R217(such as R217Q or R217E) is shifted to more negative membrane potentials [29,32,33]. X-ray crystal structure of the R217E VSD of Ci-VSP was also solved by Perozo’s group [29]. Comparison of the structures of wild-type Ci-VSD (corresponding to the resting state) and R217E (corresponding to the activated state) provides a diagram of the transition occurring upon a change in membrane potential [29]. This comparison suggests that an upward screw motion of S4 for one helix turn (60 degree rotation and 5 Å translation) takes place during the VSD’s transition from the resting to the activated state. This predicted motion corresponds to the smallest value within the 5 Å to 15 Å range, which has been inferred for the motion of the VSD within voltage-gated ion channels [34]. However, the kinetics of the motion of the charge [14] and of a fluorescent tag attached to VSD [33] suggests there are intermediate states during the transition to VSD activation. This makes it possible that the structure of the wild-type protein represents a partially activated state, not the resting state, and that the observed 5-Å movement could be an underestimation. Alternatively, the motion of the VSD of VSP may be truly smaller than that of the VSD of voltage-gated ion channels. But despite several structural differences between VSP and voltage-gated ion channels, the basic mechanisms of the structural changes are similar between Ci-VSP and other VSD-containing proteins. In fact, transfer of the VSD of VSP to a viral potassium channel pore (corresponding to the pore domain of voltage-gated potassium channel) forms a protein exhibiting the activities of a voltage-gated potassium channel [35].
Relationship between VSD motion and phosphatase activity in VSP
The quantitative relationship between the motion of its VSD and its enzyme activity provides important clues to understanding the mechanisms underlying the protein’s functionality. VSD motion can be quantified electrophysiologically by detecting transient currents derived from the relative motions of positively charged amino acid residues in the transmembrane segment elicited by a change in membrane voltage [14,30]. The method used is similar to that established for measuring the gating currents of voltage-gated ion channels [36]. These transient VSP currents are often called “sensing currents,” since there is no pore to gate in VSP. An alternative method for quantifying the motion of the VSD is voltage clamp fluorometry (VCF) using an environment-sensitive fluorophore attached to a mutated cysteine at an external water-accessible site that is expected to translocate between hydrophobic and hydrophilic environments [37]. Studies of sensing currents [14,38] and VCF [33] both suggest that there are multiple transition steps during activation of VSD. On the other hand, quantification of the readout of the VSP protein function within cells is less straightforward than in voltage-gated ion channels, where the effector function (ion permeation) can be measured electrophysiologically as ionic currents. In live cells, the VSP phosphatase activity has been indirectly estimated by measuring the levels of the phosphoinositides that serve as the enzyme’s substrates. These measurements can be done using a heterologously expressed GFP-based fluorescent phosphoinositide indicator [25,39–42] or phosphoinositide-sensitive ion channels [14,25,42–45]. These studies done in heterologous expression systems such as Xenopus oocyte or HEK293 cells showed that the phosphatase activity becomes more robust as membrane potential increases. Quantitative comparison between the VSD motion and enzyme activity indicates the enzyme activity is linearly related to the motion of the VSD [38,46]. Phosphatase activity is most likely graded depending on the extent of the VSD activation, as evidenced by analysis of a voltage sensor-trapped mutant of zebrafish VSP (Dr-VSP) [38]. This mutant exhibits a Q-V relationship with two Boltzmann components over a distinct voltage range. With this mutant, enzymatic activity gradually increased over a voltage range from the resting state of the VSD to an intermediate activated state. It then increased further as the VSD moved from the intermediate state to a fully activated state. This indicates that the enzyme can be activated upon partial activation of VSD and then tuned to a greater magnitude upon full activation of VSD [38].
Critical elements for coupling between VSD and enzyme
The coupling between the VSD and enzyme is known to involve three regions: (1) a linker region between S4 and PD (from S236 to Q256 in Ci-VSP), (2) the “gating loop” in the PD (aa 398 to 413 in Ci-VSP), and (3) the C2 domain.
(1) Cytoplasmic linker segment downstream of S4 (VSD-PD linker)
A linker segment about 20 amino acids in length connects S4 and the PD. This linker segment consists of two parts: a N-terminal region not conserved in PTEN (S236–S244 in Ci-VSP) and a C-terminal region (R245-Q256 in Ci-VSP) containing multiple basic residues with high similarity to the phosphoinositide binding motif (PBM) of PTEN. Deletion of part of the linker or mutation of the basic residues in the C-terminal region of the linker markedly reduces coupling [14,43], which highlights the critical role of the linker in coupling. In the C-terminal region of the linker (PBM-like region), there are three consecutive basic residues (K252, R253 and R254 in Ci-VSP), which are highly conserved among VSP orthologs. Moreover, each of these three residues appears to have a different function: mutation of K252 in the full-length Ci-VSP protein completely eliminated coupling [44,47], whereas some coupling persisted after mutation of R253 or R254. It is noteworthy that the contributions of these basic residues to coupling are not completely separate from their contribution to the enzyme activity itself, as mutation of these basic residues in a VSP mutant lacking the VSD reduces its phosphatase activity in in vitro phosphatase assays [47].
It has been claimed that via its multiple basic residues, the PBM in PTEN interacts with phosphoinositides [48–50], and they are known to be essential for PTEN’s phosphatase activity. Like in PTEN, the cluster of basic residues in the C-terminal region of the linker in Ci-VSP is thought to interact with phosphoinositides [47]. This is supported by the finding that forced depletion of PtdIns(4,5)P2 induced by activation of the Gq-PLC cascade by a heterologously coexpressed serotonin receptor alters the coupling between the enzyme and VSD in Ci-VSP [47]. Molecular dynamics simulations showed that the positively charged residues R245, R246, R254, R256 and K257 of the PBM-like region directly interact with PtdIns(4,5)P2 via electrostatic interaction [51]. Another molecular dynamics simulation using a coarse grained method with membrane lipids and the isolated structure of the cytoplasmic region of Ci-VSP indicated that basic residues in the VSD-PD linker segment interact with phosphoinositides [52].
Structural information about the VSD-PD linker segment has been obtained from two X-ray crystallography studies. The S236–S244 segment was solved as a part of the X-ray crystal structure (4G80) of Ci-VSD, which does not contain the PD or C2 domain. This region assumed an α helical structure. In addition, within the X-ray crystal structure (3VOD) of the cytoplasmic enzyme region of Ci-VSP, the I248-K252 segment was also α helix. In a separate study entailing systematic scanning with cysteine replacement in the region extending from M240 to K257, reduction of coupling was detected with relatively regular periodicity, which is consistent with the helical nature of the linker [44]. Further, the crystal structure of the cytoplasmic enzyme region containing the PBM-like region of the linker [53] showed a salt bridge from R253 or K252 to D400 within the PD (which is called the “gating loop” as described below). This indicates that the PBM-like region directly interacts with the PD, and that the linker region includes sites that interact with phosphoinositide and with the PD, thereby providing a key node for interaction with both the membrane and PD.
(2) Gating loop in the phosphatase domain and its coupling with the VSD-PD linker
Within the PD of VSP, there is a region that differs substantially from PTEN. The region extending from amino acid 398 to 413 of Ci-VSP is not conserved in PTEN, and the corresponding segment in PTEN is shorter by five residues and is called the TI loop. It has been proposed that this loop region in Ci-VSP is called the “gating loop” [53]. Comparison of the crystal structures of the cytoplasmic region of Ci-VSP in combination with mutagenesis studies suggested a model in which the gating loop moves with VSD activation, which alters the structure of the substrate binding pocket [53]. These X-ray structures indicate the gating loop is coupled to the VSD-PD linker: K252 and R253 in the linker interact with D400 of the gating loop and G365 comprising the backbone carbonyl of the active site. Notably, K252 and R253 exchange positions between the substrate bound and unbound forms. Based on these findings, Liu et al. proposed a model in which the motion of the VSD induces a structural change in the VSD-PD linker, which in turn induces a large conformational change in the gating loop, leading to an alteration in substrate binding to the active site [53].
(3) C2 domain
The C2 domain is not directly involved in substrate binding or catalysis in PTEN. Instead, the C2 domain in PTEN plays a key role in mediating the association between the whole protein and the membrane [54–56]. In VSP, by contrast, part of the C2 domain is involved in regulating the phosphatase activity itself. Y522 of Ci-VSP is located in the loop region facing the PD and directly interacts with H332 of the PD. Cells expressing the Ci-VSP Y522A mutant exhibit greater accumulation of PtdIns(3,4)P2 than the wild-type protein. This suggests that dephosphorylation of PtdIns(3,4)P2 into PtdIns(4)P is attenuated in Y522A mutant and that Y522 is important for 3-phosphate enzyme activity [53]. Positively charged residues in a loop region of PTEN, called the C-α2 loop, are known to mediate membrane association. Four positive charges are present in the C-α2 loop of Ci-VSP. Canceling these positive charges (quadruple C2 domain mutant) induces a 10-mV shift in the voltage-dependence of the motion of the VSD. An even more remarkable shift in voltage dependence was seen in phosphatase activity when the level of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 were examined using two phosphoinositide reporters: the pleckstrin homology (PH) domain derived from tandem PH domain-containing protein 1 (TAPP1) fused with GFP to sense PtdIns(3,4)P2 and the PH domain derived from PLC-δ fused with GFP to sense PtdIns(4,5)P2 [57]. The Ci-VSP quadruple C2 domain mutant exhibited the distinct signaling profiles of these reporters, including a more than 50 mV shift in the TAPP1-GFP signal, suggesting that rates of the subreactions of the enzyme are altered in the mutant. A slight but clear shift of voltage-dependence of the VSD indicates the C2 domain influences retrograde interaction from the PD to the VSD. Altered voltage-dependence profiles of the enzyme activities of the quadruple mutant suggest that interaction between the C2 domain and membrane influence PD activity through domain-to-domain coupling.
Conformational change or distance change for substrate?
X-ray crystallographic studies of the cytoplasmic region combined with functional analysis using electrophysiology and fluorometry in Xenopus oocytes yielded a model for the regulation of phosphatase activity by the VSD [53]. In one X-ray crystal structure, the side chain of E411 was located in a hydrophobic region, slightly distant from the substrate binding pocket, thus enabling the substrate to access the active center. In other structures, however, E411 was oriented into the pocket, narrowing it. In malachite green assays of the isolated cytoplasmic region, E411 was critical for substrate specificity: mutation of E411 altered the enzyme’s preference with respect to phosphoinositide species [24]. E411 is located in the gating loop, which interacts with the VSD-PD linker. Based on these findings, it has been proposed that VSD induces gating loop movement through its interaction with the VSD-PD linker. Changes between the open and closed forms of the substrate binding pocket are then dependent on the position of E411.
An alternative model is that the enzyme activity is regulated by the change in distance between active center of the enzyme and the membrane that results from the VSD motion. Given that the substrate for this enzyme is membrane lipid, regulation of distance between the substrate binding pocket and the lipid substrate seems reasonable. In fact, PTEN accessibility to the membrane plays a central role in the enzyme’s regulation, as PTEN enzymatic activity is dependent on efficient hopping of the protein to the membrane [58,59]. The N-terminal PBM region, PD and C2 domain of PTEN are all able to bind to the membrane. The C-terminal regulatory region of PTEN competitively binds to the PD and C2 domain, thus preventing their interaction with the membrane [23,60]. Phosphorylation of the C-terminal regulatory region causes a structural change, leading to its dissociation from the PD and C2 domain, and facilitates association of the whole protein with the membrane.
The two models summarized above largely differ from each other with respect to whether structural changes take place locally (VSD-PD linker and gating loop in the first model) or extensively to the entire cytoplasmic region (for distance change toward membrane in the second model). Information regarding voltage-dependent structural changes in the cytoplasmic region is expected to be important for discriminating between these models.
Local changes in protein structure under the cell membrane reported through genetic incorporation of fluorescent unnatural amino acid (fUAA)
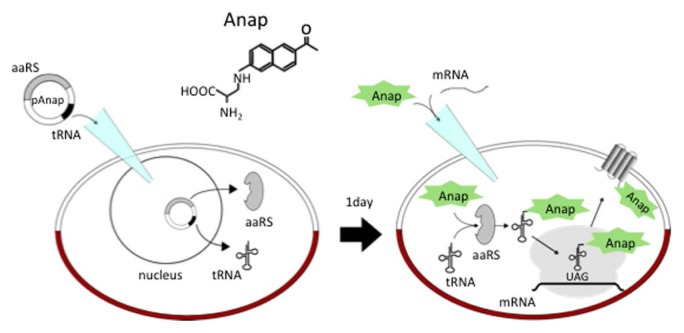
Examining dynamic changes in the local structure of the cytoplasmic region of VSP in live cells will help to understand the mechanisms underlying VSD-dependent regulation of enzymatic activity. Toward this aim, a specific site in the cytoplasmic region would be labeled such that fine structural changes could be monitored in live cells. For this purpose, however, the commonly used GFP-based fluorescent protein tags and Halo-tags are not suitable because these bulky tags would likely perturb the local structural changes or protein function. We therefore recently applied a method of genetic incorporation of Anap (3-(6-acetylnaphthalen-2-ylamino)-2-aminopropanoic acid), a fluorescent unnatural amino acid (fUAA) [61]. Anap is a fluorescent amino acid derivative of the environment-sensitive molecule Prodan [62]. Anap can be genetically incorporated into any given protein in a heterologous expression system using an orthogonal pair of aminoacyl tRNA synthetase and amber suppressor tRNA (Fig. 3), as developed in a series of pioneering studies by Dr. Schultz’s group [63–65]. Because Anap is small, it is expected that its incorporation would not perturb protein function. A plasmid carrying cDNAs encoding both amber suppressor tRNA and engineered aminoacyl tRNA synthetase was microinjected into the nucleus of a Xenopus oocyte followed by cytoplasmic introduction of cRNA encoding Ci-VSP harboring a mutation for the TAG Anap codon (Fig. 3) [66].
Figure 3.
Scheme for genetic incorporation of fUAA and its heterologous expression.
The plasmid, pAnap, carries both Anap-specific amino acyl tRNA synthetase and amber suppressor tRNA, which recognizes the TAG stop codon. Anap can be incorporated into the TAG site of the polypeptide.
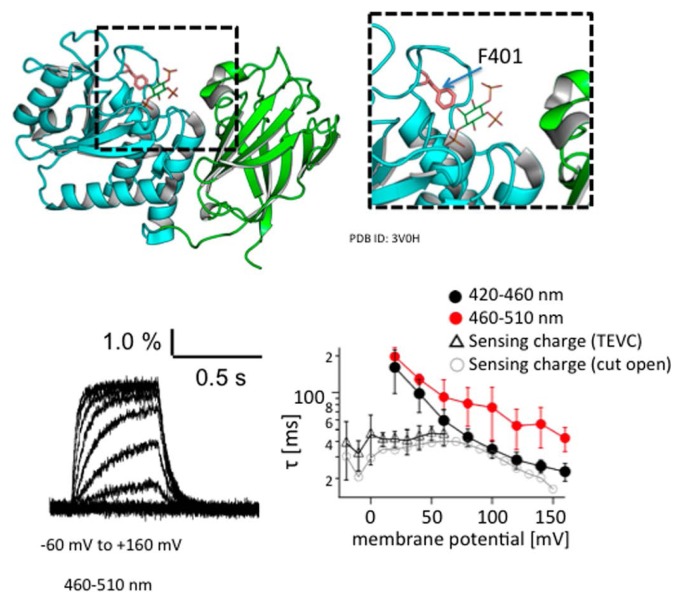
Figure 4 shows the change of Anap fluorescence at F401 within the gating loop elicited by changes in the membrane potential of a single cell in response to depolarizing steps from –60 mV to 160 mV in 20-mV increments [66]. To avoid a potential effect on Anap fluorescence of altered phosphoinositide levels due to intrinsic Ci-VSP phosphatase activity, a mutation was introduced at C363 in the enzyme domain, which is highly conserved in VSPs and other related phosphatases. This construct F401Anap/C363S showed clear voltage-dependent changes in fluorescence. Anap was also incorporated into other sites in the gating loop, including D400, F407 and Q408, which also showed voltage-dependent changes of fluorescence. These findings are consistent with the idea that the motion of the gating loop plays a central role in the regulation of VSP enzyme activity.
Figure 4.
Voltage-dependent changes of Anap fluorescence introduced at F401 of the Ci-VSP phosphatase domain (PD).
Ci-VSP with F401Anap was expressed in a Xenopus oocyte, and fluorescence through a 460–510 nm wavelength window was measured using a photomultiplier. F401 is located in the gating loop in the Ci-VSP PD (shown in pink in the top picture). Blue and green indicate PD and C2 domain, respectively. Voltage traces obtained by depolarization from −60 mV to 160 mV in 20-mV increments are superimposed. As membrane potential increased, so did the speed of the fluorescence rise. The graph shows a plot of the time constant of the rising phase against membrane potential. Data obtained with two emission filters, 420–460 nm and 460–510 nm, are indicated. Triangles and circles denote the time constant of the VSD charge motion, as measured with two-electrode voltage clamp (triangles) or the cut-open oocyte method (circles). Figures are adapted from [66].
Coupling between the C2 domain and PD suggested by results of fUAA studies
Anap was also introduced into the C2 domain to assess its voltage-dependent structural changes. Robust voltage-dependent fluorescence changes were observed at multiple positions within the C2 domain, including the 515 loop (from S513 to R520, corresponding to the CBR3 loop of PTEN) and Cα2 loop containing multiple basic residues, K553, K555 or K558, indicating the motion was not confined to the gating loop of the PD, but extended to the C2 domain [66]. Moreover, when the latency prior the start of the fluorescence change at several sites within the gating loop and C2 domain was compared, no significant difference in latency was detected, suggesting that multiple regions of the cytoplasmic enzyme change their conformation nearly simultaneously.
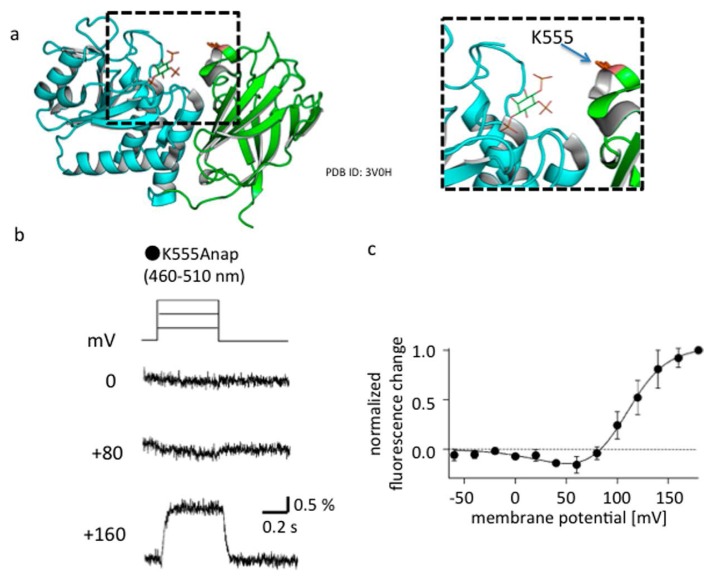
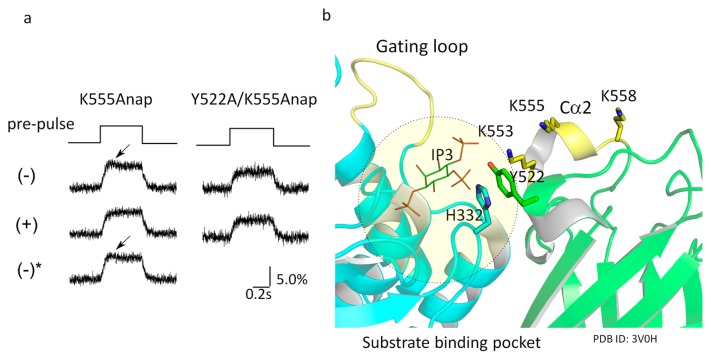
Measurements of Anap fluorescence from the C2 domain also brought two other important results [66]. First, when Anap replaced K520, K553, K555 or K558, the voltage dependence of the fluorescence changes was biphasic: fluorescence decreased at 80 mV but increased with greater depolarizations (Fig. 5 for the example of K555Anap). This suggests the conformational change in the cytoplasmic region differs at different membrane potentials. This will be discussed later. Second, the motion of the C2 domain was sensitive to availability of substrate, most likely in the substrate-binding pocket (Fig. 6). When Anap replaced K555, one of the basic residues of the Cα2 loop of the C2 domain, the kinetics of the Anap fluorescence during a step to 160 mV was dependent on the pulse protocol. When a depolarizing test step was applied with long pulse-to-pulse interval (60 s), the signal showed a biphasic pattern, first a rise and then a decline (Fig. 6a, K555Anap, (–)). By contrast, when a depolarizing conditioning pulse (3 s to 100 mV) preceded the test pulse in the same cell, the early transient phase of the fluorescence change was not observed (Fig. 6a, K555Anap, (+)). This depolarizing conditioning pulse has previously been shown to induce depletion of PtdIns(4,5)P2 by Ci-VSP, indicating that the sensitivity of the pattern of Anap fluorescence to the pulse protocol reflects the sensitivity of local structural changes due to VSD motion to availability of PtdIns(4,5)P2. The Cα2 loop is located close to Y522 in the C2 domain, which faces the substrate binding pocket in the PD through interaction with H332. Because K555 forms hydrogen bonds with neighboring charged residues, it may indirectly interact with the substrate binding pocket. Y522 was mutated to alanine, which weakens its interaction with H332 in the PD. In this mutant, the pulse protocol-dependent changes in Anap fluorescence was not observed [66] (Fig. 6a, Y552A/K555Anap).
Figure 5.
Biphasic profile of fluorescence-voltage relationship of Anap introduced at K555
(a) The site of Anap incorporation (K555 of C2 domain) is shown in orange. Blue and green indicate PD and C2 domain, respectively. (b) Voltage-dependent changes in the fluorescence of Anap introduced at K555. Note that the signal decreases at 80 mV but increases at 160 mV. The voltage protocol is at the top. The holding potential was −60 mV. Depolarizing steps were applied for 1.2 s. (c) The fluorescence-voltage relationship of K555Anap shows a biphasic pattern.
Figure 6.
Evidence that movement of the C2 domain is sensitive to the availability of substrate in the binding pocket.
(a) Voltage-dependent changes in Anap fluorescence evoked by 160-mV depolarizations for 1.2 s. All measurements were from the same Xenopus oocyte. Anap was introduced at K555. (−) indicates that there was no conditioning prepulse, only with test pulse. (+) indicates that a depolarizing conditioning pulse to 100 mV was applied for 3 s prior to the test pulse to 160 mV. Depolarization to 100 mV for 3 s depletes most PtdIns(4,5)P2 from Xenopus oocytes due to the enzymatic activity of Ci-VSP. (−)* indicates the trace was obtained after polarizing the membrane to −60 mV for 60 s. Note that early hump (arrow) is only seen when the conditioning pulse was not applied. Data are adapted from [66]. (b) Interface between the C-α2 loop region (yellow ribbon) of the C2 domain and the substrate binding pocket (dotted circle) in the PD. The Anap site, K555, interacts with K553 and Y522. Y522 interacts with H332, which is located near the substrate. The structure is from information deposited as 3V0H in the PDB. This X-ray crystal structure contains myo-inositol 1,4,5-trisphosphate (InsP3) (red) in the binding pocket.
Changes in membrane potential produce little change in the distance between the cytoplasmic region and membrane
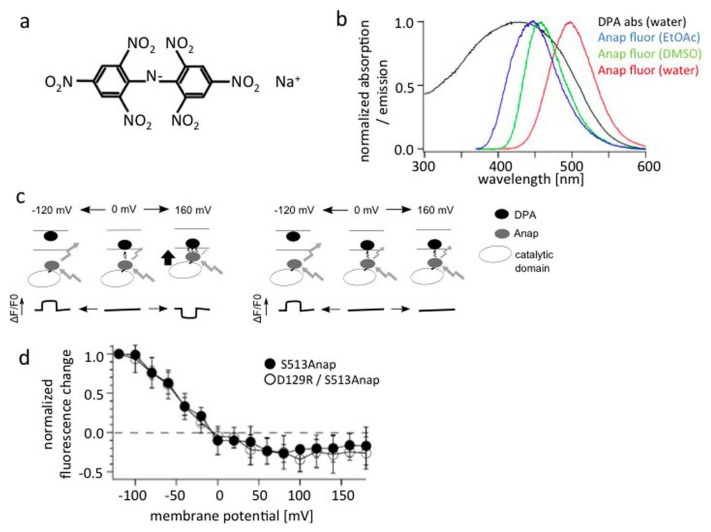
Dipicrylamine (DPA) is a membrane associated charged molecule (Fig. 7a) that absorbs fluorescence with a peak wavelength of 420 nm. Because its absorption spectrum covers the emission spectrum of Anap (Fig. 7b), DPA can serve as a FRET acceptor for Anap. This provides a unique opportunity to use FRET measurements to estimate the distance between the membrane (labeled by DPA) and the VSP cytoplasmic region (labeled by Anap). One complication with using DPA as a FRET acceptor is that DPA is redistributed between the two membrane leaflets depending on membrane potential [67]. In the experimental setup, if the membrane potential was hyperpolarized, an increase in Anap fluorescence was expected, since DPA travels to the outer leaflet. If the cytoplasmic region comes closer to the membrane upon depolarization, Anap fluorescence will be absorbed by DPA, so the signal is expected to decrease. However, if a voltage change does not lead to movement of the cytoplasmic region of VSP, the Anap signal may not change upon membrane depolarization (Fig. 7c). Because Anap fluorescence is being used as the donor fluorescence for FRET in this setup, L518 and S513, at which Anap fluorescence does not change significantly with movement of the VSD, were chosen as the Anap sites.
Figure 7.
FRET measurements using DPA and Anap show little change in distance between the membrane and the PD.
(a) Structure of DPA. (b) Absorbance spectrum of DPA (black) and emission spectrum of Anap (other colors). The latter is covered by the former, suggesting DPA serves as a FRET acceptor for Anap emission. (c) The idea behind the use of FRET measurements to detect movement of the Ci-VSP cytoplasmic region toward the membrane. Membrane is labeled with DPA (black ellipse), and the Ci-VSP cytoplasmic region is labeled with Anap (shaded ellipse). DPA is known to translocate between the two membrane leaflets in a voltage-dependent manner. Voltage-dependent changes in the distance between the DPA and Anap are expected to be reported by a decrease in Anap fluorescence upon membrane depolarization (e.g., by a step pulse to 160 mV). (d) FRET measurements with Anap incorporation at S513. This site is known to remain immobile during changes in membrane potential, as judged by the absence of significant changes in the fluorescence of Anap incorporated at the site. The slight decrease of Anap fluorescence most likely reflects voltage-dependent movement of DPA within the membrane. There is no difference between S513Anap and a voltage sensor-defective double mutant (D129R/S513Anap). b, c, d are adapted from [66].
Figure 7d shows the results of FRET assays for a protein with S513Anap. After a voltage step to −120 mV from the holding potential of 0 mV, an increase in fluorescence was observed reflecting the intrinsic voltage-sensitive motion of DPA to outer leaflet. Upon depolarization to 160 mV, some decrease of fluorescence was observed. This decrease at 160 mV is most likely due to unsaturated motion of DPA at high membrane depolarization, since similar weak reductions were seen upon depolarization to 160 mV in the background of the voltage sensor-defective D129R mutant. These findings indicate that voltage sensor activation does not lead to a significant change in the distance between the membrane and cytoplasmic region. This is consistent with stable association of the whole cytoplasmic region of Ci-VSP with the membrane as shown in a molecular dynamics simulation [52].
Coupling mechanisms implicated by VCF study using fUAA
The results of Anap studies [66] are consistent with the gating loop playing a central role in the regulation of VSP enzyme activity [53]. On the other hand, no large motion was detected in FRET-based measurements using Anap and DPA, which rules out any model that requires a large change in the distance between the membrane and enzyme upon activation of the VSD. The results of the Anap experiment also provided new insights into the coupling mechanism. The motion of the loop regions of the C2 domain detected with the Anap signal was as rapid as that of the gating loop, at least within limited time resolution of the signal detection under two-electrode voltage clamp. Such rapid motion of the C2 domain was unexpected, since the C2 domain is only remotely coupled to the VSD through interaction with the PD. This indicates that the whole cytoplasmic region of Ci-VSP undergoes an extensive change in structure upon activation of VSD motion, and that the motion is not restricted to the gating loop. Moreover, Anap introduced into the C2 domain enabled substrate binding or the state of catalysis to be monitored, indicating that tight coupling between the C2 domain and PD contributes to the enzyme reaction. A loop in VSP in the region corresponding to the CBR3 loop in the C2 domain of PTEN interacts with several key residues at the active center of the PD, whereas the CBR3 loop in PTEN faces toward the membrane. Mutation of Y522 is known to change substrate preference [53], and the sensitivity of Anap fluorescence from the C2 domain to substrate availability [66] (Fig. 6a) probably reflects involvement of the C2 domain at certain steps during the enzymatic dephosphorylation of phosphoinositide substrates. Therefore, the effect of VSD motion is transmitted to the entire structure of the cytoplasmic region for enzyme activation.
Recordings of Anap fluorescence at different membrane potentials from the same oocytes showed a complex profile of local structural changes (Fig. 5b, c). At four sites of Anap incorporation within the C2 domain (R520, K553, K555, K558), the fluorescence-voltage relationship showed both a decrease and an increase in fluorescence depending on the membrane potential. This indicates that local protein structures around these sites make distinct conformational changes depending on the membrane voltage. Studies have suggested that activation of the VSD in Ci-VSP involves multiple steps, not a single transition [47]. Perhaps, distinct patterns of conformational changes are coupled to particular states of the voltage sensor. Grimm and Isacoff studied Ci-VSP enzyme activity in live cells under voltage clamp using a rapid fluorescent indicator for each species of phosphoinositide [40]. As mentioned, VSP dephosphorylates three species of phosphoinositides, PtdIns(3,4,5)P3, PtdIns(3,4)P2, PtdIns(4,5)P2 (Fig. 2c). Based on measurements of phosphoinositide dynamics at different membrane potentials with the wild-type protein and VSD mutants, it was proposed that distinct activated states of the VSD induce different states of enzyme activity biased toward PIP3 or PIP2 (PtdIns(3,4)P2 and PtdIns(4,5)P2). With mild depolarization, VSP would preferentially dephosphorylate PtdIns(3,4,5)P3; with greater depolarization, however, VSP would exhibit robust activity toward PtdIns(3,4)P2 and PtdIns(4,5)P2. The complex patterns of Anap fluorescence changes [66] in the constructs, including K555 Anap, may reflect these multiple states of the enzyme region with distinct substrate preferences. Recently Keum et al. performed a careful analysis entailing fluorescence-based measurements of the level of each species of phosphoinositide and mathematical modeling [41]. In this study, the complex changes in phosphoinositide at different voltage levels observed by Grimm et al. [40] with both of the wild-type protein and VSD mutants were recapitulated by mathematical modeling with shared voltage dependence among the subreactions of the enzyme with the different phosphoinositide substrates [41].
Whether substrate preference varies depending on membrane potential or not, recent findings with voltage-gated ion channels suggest it is reasonable to think the enzyme can assume different structures depending on the activation state of the VSD [68,69]. In most voltage-gated ion channels such as voltage-gated sodium channels or voltage-gated potassium channels, full activation of multiple VSDs is necessary for pore opening. But it was recently shown that the KCNQ1 potassium channel is able to conduct potassium ions upon even partial activation of the VSD [68]. Partially activated and fully activated states of individual VSDs are coupled to distinct opening states of the PGD [69]. It would be interesting to know how the basic principle of coupling particular effector conditions to different states of the VSD is conserved between these two distinct classes of proteins harboring VSDs.
Perspectives
Since the initial characterization of the molecular properties of Ci-VSP [14], structural information about the individual domains comprising VSP, as well as the findings of biophysical studies, have provided insight into the mechanisms by which these domains are functionally coupled. Genetic methods using fUAA are versatile tools for detecting structural changes in the cytoplasmic region and a recent application of this method to Ci-VSP [66] has enabled us to gain novel insight into the dynamics of the enzyme region. It is anticipated that more information on the physicochemical basis for the observed alterations in Anap fluorescence within proteins will further expand usefulness of this method. Greater understanding of the coupling mechanisms operating within VSP will lead to better understanding of the principles of domain-to-domain coupling shared by many proteins and facilitate the engineering of molecules (e.g., a voltage reporter for imaging electrical activity) for basic research and other purposes.
Significance.
Neuron, muscle cell, endocrine cell and sperm utilize signals of membrane potential. Such signals depend on activities of diverse types of voltage-gated ion channels. Voltage-gated ion channels share common structures: voltage sensor domain and pore-gate domain. Voltage-sensing phosphatase (VSP) does not contain pore-gate domain but instead the cytoplasmic enzyme region. The enzyme consists of the phosphatase domain and C2 domain and dephosphorylate phosphoinositides, like tumor suppressor phosphatase, PTEN. Conformational change of voltage sensor domain induces structural change of the enzyme. Recent studies have revealed details of relationships between voltage sensor domain and enzyme in VSP.
Acknowledgements
We thank Ms. Yuka Jinno for her help in conducting experiments related to this work, Dr. Fumihito Ono at Osaka Medical College for helpful support, Drs. Hirotaka Narita and Atsushi Nakagawa of the Institute for Protein Research at Osaka University, and members of Okamura lab of for helpful discussion. This work was supported by Grants-in-Aids from Ministry of Education, Culture, Sports, Science, and Technology (MEXT) (15H05901, 16H02617, 25253016 to Y.O.) and Core Research for Evolutional Science and Technology, Japan Science and Technology Agency (CREST, JST) (JPMJCR14M3 to Y.O.).
Abbreviations
- VSD
voltage-sensor domain
- PGD
pore-gate domain
- VSP
voltage-sensing phosphatase
- PD
phosphatase domain
- Cav channel
voltage-gated calcium channel
- Kv channel
voltage-gated potassium channel
- PtdIns(3,4,5)P3
phosphatidylinositol-3,4,5-trisphosphate
- PtdIns(4,5)P2
phosphatidylinositol-4,5-bisphosphate
- fUAA
fluorescent unnatural amino acid
- PBM
phosphoinositide binding motif
- PH domain
pleckstrin homology domain; 3-(6-acetylnaphthalen-2-ylamino)-2-aminopropanoic acid (Anap)
- PTEN
phosphatase and tensin homolog deleted on chromosome ten
Footnotes
Conflict of Interest
The authors declare no conflict of interests.
Author contributions
S.S., M.M., A.K. and Y.O. wrote the manuscript.
References
- 1.Hille B. Ion channels of excitable membranes. Sinauer. 2001 [Google Scholar]
- 2.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 3.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Yan Z, Li Z, Qian X, Lu S, Dong M, et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 A resolution. Nature. 2016;537:191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
- 5.Tao X, Hite RK, MacKinnon R. Cryo-EM structure of the open high-conductance Ca2+-activated K+ channel. Nature. 2016;541:46–51. doi: 10.1038/nature20608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whicher JR, MacKinnon R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CH, MacKinnon R. Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell. 2017;168:111–120.e11. doi: 10.1016/j.cell.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura Y, Fujiwara Y, Sakata S. Gating mechanisms of voltage-gated proton channels. Annu Rev Biochem. 2015;84:685–709. doi: 10.1146/annurev-biochem-060614-034307. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara Y, Kurokawa T, Takeshita K, Kobayashi M, Okochi Y, Nakagawa A, et al. The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H+ channel Hv1. Nat Commun. 2012;3:816. doi: 10.1038/ncomms1823. [DOI] [PubMed] [Google Scholar]
- 12.Okuda H, Yonezawa Y, Takano Y, Okamura Y, Fujiwara Y. Direct Interaction between the Voltage Sensors Produces Cooperative Sustained Deactivation in Voltage-gated H+ Channel Dimers. J Biol Chem. 2016;291:5935–5947. doi: 10.1074/jbc.M115.666834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Shen R, Treger JS, Wanderling SS, Milewski W, Siwowska K, et al. Resting state of the human proton channel dimer in a lipid bilayer. Proc Nat Acad Sci USA. 2015;112:E5926–5935. doi: 10.1073/pnas.1515043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 15.Hossain MI, Iwasaki H, Okochi Y, Chahine M, Higashijima S, Nagayama K, et al. Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J Biol Chem. 2008;283:18248–18259. doi: 10.1074/jbc.M706184200. [DOI] [PubMed] [Google Scholar]
- 16.Ratzan WJ, Evsikov AV, Okamura Y, Jaffe LA. Voltage sensitive phosphoinositide phosphatases of Xenopus: their tissue distribution and voltage dependence. J Cell Physiol. 2011;226:2740–2746. doi: 10.1002/jcp.22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Kurokawa T, Taira I, Aoki N, Sakata S, Okamura Y, et al. Potential role of voltage-sensing phosphatases in regulation of cell structure through the production of PI(3,4)P2. J Cell Physiol. 2014;229:422–433. doi: 10.1002/jcp.24463. [DOI] [PubMed] [Google Scholar]
- 18.Rosasco MG, Gordon SE, Bajjalieh SM. Characterization of the Functional Domains of a Mammalian Voltage-Sensitive Phosphatase. Biophys J. 2015;109:2480–2491. doi: 10.1016/j.bpj.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halaszovich CR, Leitner MG, Mavrantoni A, Le A, Frezza L, Feuer A, et al. A human phospholipid phosphatase activated by a transmembrane control module. J Lipid Res. 2012;53:2266–2274. doi: 10.1194/jlr.M026021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guipponi M, Tapparel C, Jousson O, Scamuffa N, Mas C, Rossier C, et al. The murine orthologue of the Golgi-localized TPTE protein provides clues to the evolutionary history of the human TPTE gene family. Hum Genet. 2001;109:569–575. doi: 10.1007/s004390100607. [DOI] [PubMed] [Google Scholar]
- 21.Walker SM, Downes CP, Leslie NR. TPIP: a novel phosphoinositide 3-phosphatase. Biochem J. 2001;360:277–283. doi: 10.1042/0264-6021:3600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Dowbenko D, Pisabarro MT, Dillard-Telm L, Koeppen H, Lasky LA. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J Biol Chem. 2001;276:21745–21753. doi: 10.1074/jbc.M101480200. [DOI] [PubMed] [Google Scholar]
- 23.Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Nat Acad Sci USA. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda M, Takeshita K, Kurokawa T, Sakata S, Suzuki M, Yamashita E, et al. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J Biol Chem. 2011;286:23368–23377. doi: 10.1074/jbc.M110.214361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata Y, Okamura Y. Depolarization activates the phosphoinositide phosphatase Ci-VSP, as detected in Xenopus oocytes coexpressing sensors of PIP2. J Physiol. 2007;583:875–889. doi: 10.1113/jphysiol.2007.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki H, Murata Y, Kim Y, Hossain MI, Worby CA, Dixon JE, et al. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. Proc Nat Acad Sci USA. 2008;105:7970–7975. doi: 10.1073/pnas.0803936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurokawa T, Takasuga S, Sakata S, Yamaguchi S, Horie S, Homma KJ, et al. 3′ Phosphatase activity toward phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] by voltage-sensing phosphatase (VSP) Proc Nat Acad Sci USA. 2012;109:10089–10094. doi: 10.1073/pnas.1203799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacroix J, Halaszovich CR, Schreiber DN, Leitner MG, Bezanilla F, Oliver D, et al. Controlling the activity of a phosphatase and tensin homolog (PTEN) by membrane potential. J Biol Chem. 2011;286:17945–17953. doi: 10.1074/jbc.M110.201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villalba-Galea CA, Frezza L, Sandtner W, Bezanilla F. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J Gen Physiol. 2013;142:543–555. doi: 10.1085/jgp.201310993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsutsui H, Jinno Y, Tomita A, Okamura Y. Optically detected structural change in the N-terminal region of the voltage-sensor domain. Biophys J. 2013;105:108–115. doi: 10.1016/j.bpj.2013.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akemann W, et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS ONE. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 34.Borjesson SI, Elinder F. Structure, function, and modification of the voltage sensor in voltage-gated ion channels. Cell Biochem Biophys. 2008;52:149–174. doi: 10.1007/s12013-008-9032-5. [DOI] [PubMed] [Google Scholar]
- 35.Arrigoni C, Schroeder I, Romani G, Van Etten JL, Thiel G, Moroni A. The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J Gen Physiol. 2013;141:389–395. doi: 10.1085/jgp.201210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 37.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 38.Sakata S, Okamura Y. Phosphatase activity of the voltage-sensing phosphatase, VSP, shows graded dependence on the extent of activation of the voltage sensor. J Physiol. 2014;592:899–914. doi: 10.1113/jphysiol.2013.263640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halaszovich CR, Schreiber DN, Oliver D. Ci-VSP is a depolarization-activated phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate 5′-phosphatase. J Biol Chem. 2009;284:2106–2113. doi: 10.1074/jbc.M803543200. [DOI] [PubMed] [Google Scholar]
- 40.Grimm SS, Isacoff EY. Allosteric substrate switching in a voltage-sensing lipid phosphatase. Nat Chem Biol. 2016;12:261–267. doi: 10.1038/nchembio.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keum D, Kruse M, Kim D, Hille B, Suh B. Phosphoinositide 5- and 3-phosphatase activities of a voltage-sensing phosphatase in living cells show identical voltage dependence. Proc Nat Acad Sci USA. 2016;113:E3686–3695. doi: 10.1073/pnas.1606472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falkenburger BH, Jensen JB, Hille B. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J Gen Physiol. 2010;135:99–114. doi: 10.1085/jgp.200910345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villalba-Galea CA, Miceli F, Taglialatela M, Bezanilla F. Coupling between the voltage-sensing and phosphatase domains of Ci-VSP. J Gen Physiol. 2009;134:5–14. doi: 10.1085/jgp.200910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobiger K, Utesch T, Mroginski MA, Friedrich T. Coupling of Ci-VSP modules requires a combination of structure and electrostatics within the linker. Biophys J. 2012;102:1313–1322. doi: 10.1016/j.bpj.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamura Y, Dixon JE. Voltage-sensing phosphatase: its molecular relationship with PTEN. Physiology (Bethesda) 2011;26:6–13. doi: 10.1152/physiol.00035.2010. [DOI] [PubMed] [Google Scholar]
- 46.Sakata S, Hossain MI, Okamura Y. Coupling of the phosphatase activity of Ci-VSP to its voltage sensor activity over the entire range of voltage sensitivity. J Physiol. 2011;589:2687–2705. doi: 10.1113/jphysiol.2011.208165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohout SC, Bell SC, Liu L, Xu Q, Minor DL, Jr, Isacoff EY. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat Chem Biol. 2010;6:369–375. doi: 10.1038/nchembio.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- 49.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 50.Campbell RB, Liu F, Ross AH. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 51.Hobiger K, Utesch T, Mroginski MA, Seebohm G, Friedrich T. The linker pivot in Ci-VSP: the key to unlock catalysis. PloS ONE. 2013;8:e70272. doi: 10.1371/journal.pone.0070272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalli AC, Devaney I, Sansom MS. Interactions of phosphatase and tensin homologue (PTEN) proteins with phosphatidylinositol phosphates: insights from molecular dynamics simulations of PTEN and voltage sensitive phosphatase. Biochemistry. 2014;53:1724–1732. doi: 10.1021/bi5000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, Kohout SC, Xu Q, Muller S, Kimberlin CR, Isacoff EY, et al. A glutamate switch controls voltage-sensitive phosphatase function. Nat Struct Mol Biol. 2012;19:633–641. doi: 10.1038/nsmb.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 55.Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Nat Acad Sci USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Georgescu MM, Kirsch KH, Kaloudis P, Yang H, Pavletich NP, Hanafusa H. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 2000;60:7033–7038. [PubMed] [Google Scholar]
- 57.Castle PM, Zolman KD, Kohout SC. Voltage-sensing phosphatase modulation by a C2 domain. Front Pharmacol. 2015;6:63. doi: 10.3389/fphar.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vazquez F, Matsuoka S, Sellers WR, Yanagida T, Ueda M, Devreotes PN. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc Nat Acad Sci USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yasui M, Matsuoka S, Ueda M. PTEN hopping on the cell membrane is regulated via a positively-charged C2 domain. PLoS Comput Biol. 2014;10:e1003817. doi: 10.1371/journal.pcbi.1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Odriozola L, Singh G, Hoang T, Chan AM. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem. 2007;282:23306–23315. doi: 10.1074/jbc.M611240200. [DOI] [PubMed] [Google Scholar]
- 61.Lee HS, Guo J, Lemke EA, Dimla RD, Schultz PG. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J Am Chem Soc. 2009;131:12921–12923. doi: 10.1021/ja904896s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber G, Farris FJ. Synthesis and spectral properties of a hydrophobic fluorescent probe: 6-propionyl-2-(dimethylamino) naphthalene. Biochemistry. 1979;18:3075–3078. doi: 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Xie J, Schultz PG. A genetically encoded fluorescent amino acid. J Am Chem Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 64.Fleissner MR, Brustad EM, Kalai T, Altenbach C, Cascio D, Peters FB, et al. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Nat Acad Sci USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chatterjee A, Guo J, Lee HS, Schultz PG. A genetically encoded fluorescent probe in mammalian cells. J Am Chem Soc. 2013;135:12540–12543. doi: 10.1021/ja4059553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakata S, Jinno Y, Kawanabe A, Okamura Y. Voltage-dependent motion of the catalytic region of voltage-sensing phosphatase monitored by a fluorescent amino acid. Proc Nat Acad Sci USA. 2016;113:7521–7526. doi: 10.1073/pnas.1604218113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci. 2005;8:1619–1626. doi: 10.1038/nn1558. [DOI] [PubMed] [Google Scholar]
- 68.Zaydman MA, Kasimova MA, McFarland K, Beller Z, Hou P, Kinser HE, et al. Domain-domain interactions determine the gating, permeation, pharmacology, and subunit modulation of the IKs ion channel. Elife. 2014;3:e03606. doi: 10.7554/eLife.03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cui J. Voltage-Dependent Gating: Novel Insights from KCNQ1 Channels. Biophys J. 2016;110:14–25. doi: 10.1016/j.bpj.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]