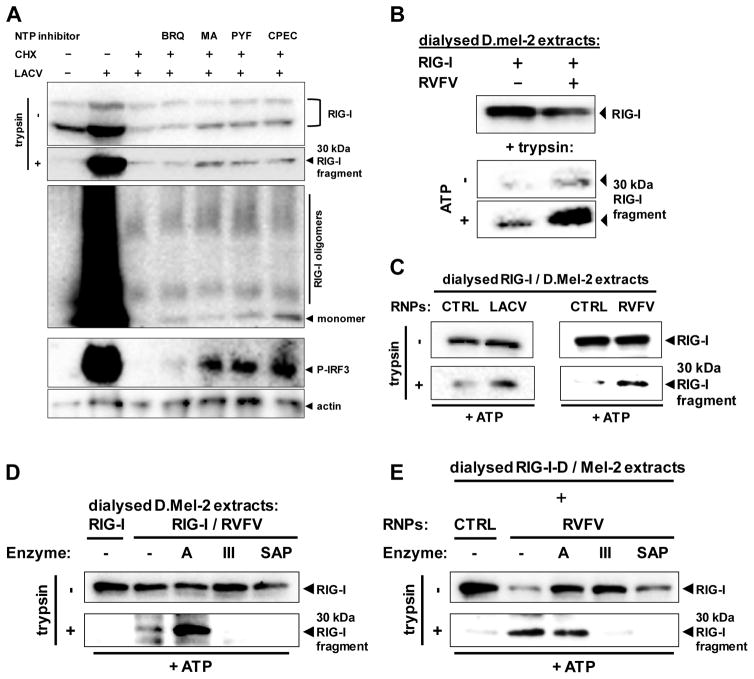
Fig. 6. Activation of RIG-I solely depends on the nucleocapsid-borne panhandle structure.
(A) Effect of NTP withdrawal on activation of RIG-I and IRF-3. Pretreated A549 cells were infected with LACVdelNSs for 5 h. Pretreatment with CHX (50 μg/ml) was for 1h, and with BRQ (10 μM, stocks dissolved in DMSO), MA (10 μM, stocks dissolved in methanol), PYF (10 μM, stocks dissolved in DMSO), or CPEC (5 μM, stocks dissolved in DMSO) for 24 h. RIG-I conformation (upper two panels), RIG-I oligomerization (upper middle panel), and IRF-3 phosphorylation (lower middle panel) were monitored. Immunoblot for actin served as loading control. (B) RIG-I activation in dialysed samples. Lysates from D.Mel-2 cells expressing RIG-I (RIG-I) or infected with RVFVΔNSs::REN (RVFV) were dialysed against PBS, mixed with each other, and incubated with or without ATP. After 1 h incubation, mixes were subjected to the RIG-I conformational switch assay. (C) RIG-I activation by purified viral nucleocapsids (RNPs). Lysates from D.Mel-2 cells expressing RIG-I were dialysed against PBS and mixed with purified nucleocapsids from particles of LACV (left panels) or RVFV (right panels). Incubation with ATP and conformational switch assay were performed as described for (B). Equivalent fractions of gradient-purified supernatants from mock-infected cells were used as negative control (CTRL). (D and E) Structural requirements for viral nucleocapsids to activate RIG-I. Lysates from RVFV-infected D.Mel-2 cells (D) or purified RVFV nucleocapsids (E) were incubated with ATP and one of the indicated enzymes, namely RNase A (A), RNase III (III), or Shrimp Alkaline Phosphatase (SAP). After mixing and incubation with dialysed lysates from RIG-I-expressing D.Mel-2 cells, the RIG-I conformational switch assay was performed. Negative controls (CTRL) were performed as described for (C). See also Figs. S17 and S18.