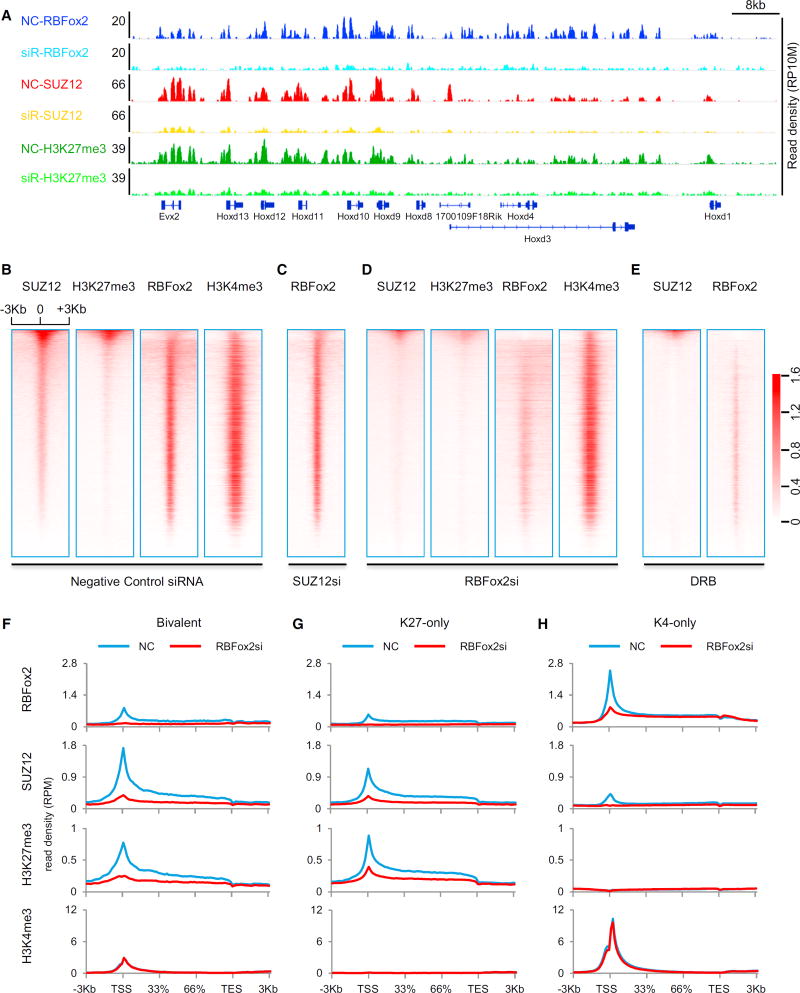
Figure 3. Functional Requirement for RBFox2 to Mediate Global PRC2 Targeting.
(A) ChIP-seq signals on a representative chromosomal segment for RBFox2, SUZ12, and H3K27me3 before and after siRNA-mediated RBFox2 knockdown (siR) in MEFs. The y axis indicates the read density per 10 million total reads (RP10M).
(B–E) Sorted ChIP-seq signals for SUZ12, H3K27me3, RBFox2, and H3K4me3 at TSS regions in control siRNA-treated (B), SUZ12 siRNA-treated (C), RBFox2 siRNA-treated (D), and DRB-treated (E) MEFs. The data were sorted according to mean SUZ12 ChIP-seq signals in control siRNA-treated MEFs. All ChIP-seq data were normalized by total reads after subtracting input signals. The numbers beside the color bar indicate the read density per million. See also Figure S3 and Table S3.
(F–H) Responses of the indicated ChIP-seq signals to RBFox2 knockdown on different gene classes (bivalent, H3K27me3 only, or H3K4me3 only) in MEFs.
See also Figure S4 and Table S4 on related functional data from C2C12 cells.