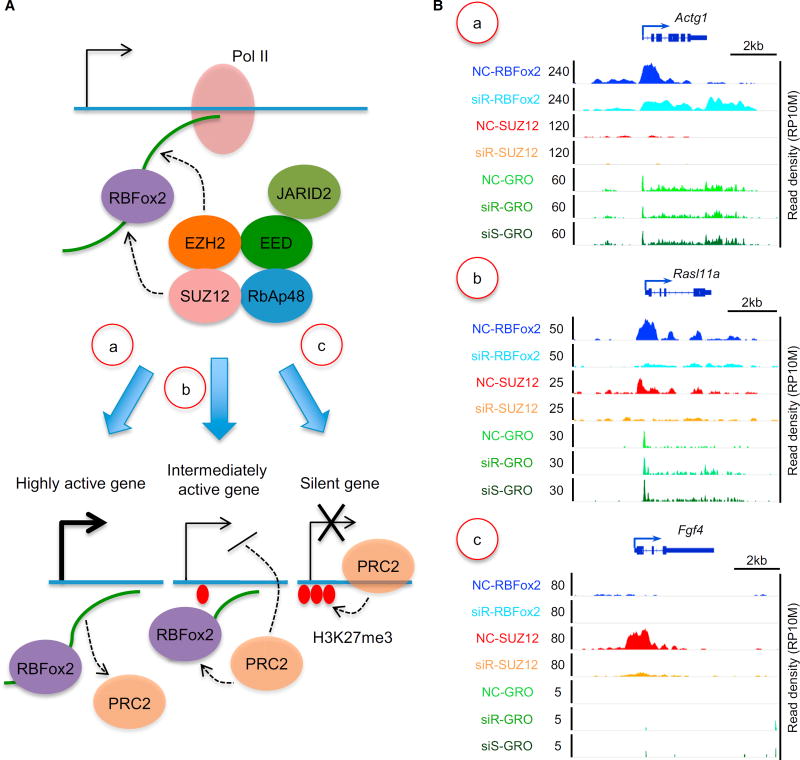
Figure 6. Proposed Model for Functional Interplays of RBFox2, PRC2, and H3K27me3 that Result in Different Gene Classes.
(A) Model for nascent RNA-mediated, RBFox2-dependent recruitment of PRC2. (a) On highly expressed genes, nascent RNA may modulate the RNA sensor function of PRC2 to cause PRC2 repulsion. (b) On modestly expressed genes, functional interplay between RBFox2 and PRC2 may dynamically regulate homeostatic gene expression, as a modest increase or decrease in nascent RNA production may cause increased or decreased PRC2 recruitment to induce feedback controls. A level of pre-deposited H3K27me3 may help stabilize recruited PRC2. (c) On nearly silent genes, interaction of PRC2 with chromatin is switched from RNA-dependent to EED-mediated interactions, leading to the amplification of the repressive signal to eventually shut down the genes. It is important to also emphasize that functional outcomes on individual genes likely depend on the contribution of transcription regulators in conjunction with specific nucleosome states.
(B) Representative genes indifferent classes based on the genomic data generated in the current study: NC, siR, and siS represent ChIP-seq or GRO-seq signals in nonspecific control siRNA-, siRBFox2-, and siSUZ12-treated MEFs, respectively. (a–c) Examples of specific gene in each class described in (A, a–c), showing RBFox2 and SUZ12 binding before and after RBFox2 knockdown, as well as nascent RNA production before and after RBFox2 or SUZ12 knockdown. NC, nonspecific siRNA; siR, siRBFox2; siS, siSUZ12.