Figure 2.
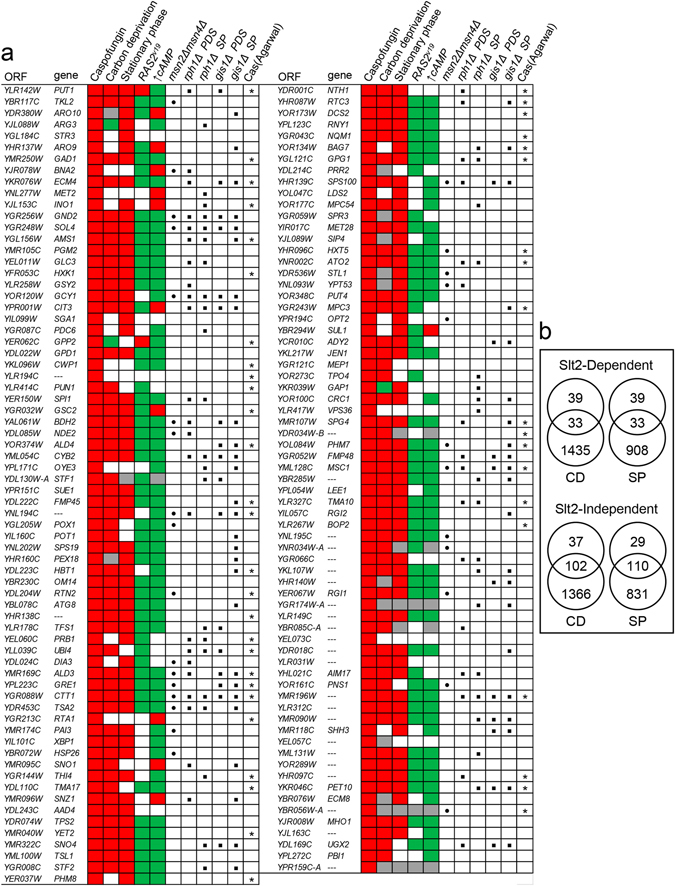
The genes induced by caspofungin independently of Slt2 are targets of the cAMP/PKA pathway. (a) Open reading frames whose transcripts were induced at least twofold in experiments of DNA microarrays in the wild-type BY4741 strain, independently of Slt2, after 2 h of CAS treatment, are shown and compared to different transcriptional genome-wide datasets using the MEV (Multiexperiment Viewer) Version 4.9 software from TIGR67: Carbon deprivation (YNB lacking D-glucose for 2 h)68, stationary phase (YPD medium for 2 days)20, RAS2 val19 (cells expressing a constitutively active allele of RAS2) (taken from GEO database GSE8805), and high levels of cAMP (medium including 1 mM cAMP)69. Green and red boxes indicate those genes repressed and induced at least twofold respectively in each condition. Gray boxes denote missing values. Those genes that were found to be dependent on Msn2/Msn4 for activation by CAS from DNA microarrays are labeled with a black dot. Black squares indicate those genes previously reported to be dependent on Rph1 and Gis1 transcription factors after diauxic shift (PDS) (wild-type strain grown in glucose depletion for 9 h) and/or stationary phase (SP) (wild-type strain grown in YPD for 3 days)29. Those genes previously described upregulated in the study of Agarwal et al.12 are marked with an asterisk. (b) Comparison of the overlap of Slt2-dependent or independent genes with those induced by stationary phase (SP) or carbon deprivation (CD).
