Abstract
In Plantae, the Calvin–Benson–Bassham (CBB) cycle is highly regulated and most of its enzymes have been thoroughly studied. Since diatoms arose as a result of secondary endosymbiosis with one or more Plantae ancestors, their precise evolutionary history is enigmatic and complex resulting in biochemical variations on the original CBB cycle theme. The Rubisco Michaelis constant for CO2 is higher in diatoms than land plants and the nuclear-encoded Rubisco activase in Plantae is replaced by an analogous chloroplast-encoded CbbX (Calvin–Benson–Bassham protein X) in diatoms. In the CBB cycle reduction phase, phosphoglycerate kinase in diatoms is redox-regulated and similar to that in red algae; however, glyceraldehyde phosphate dehydrogenase (GAPDH) is not redox-regulated, unlike in Plantae. The phosphoribulokinase (PRK)-GAPDH-CP12 complex found in many photosynthetic organisms has not yet been found in diatoms, but a ferredoxin-NADP reductase (FNR)-GAPDH-CP12 complex has been found in one species. In the CBB cycle regeneration phase, sedoheptulose 1,7-bisphosphatase and PRK are not redox-regulated in diatoms, unlike in Plantae. Regulation at the transcriptional level seems to be important in diatoms. CBB cycle enzyme properties appear to be variable among diatoms, but this view relies on results from a few model species: a greater range of diatoms need to be studied to test this.
This article is part of the themed issue ‘The peculiar carbon metabolism in diatoms’.
Keywords: GAPDH, PGK, PRK, redox-regulation, Rubisco
1. Introduction
Chloroplasts are pivotal to the most fundamental biochemical reactions upon which photosynthetic eukaryotes depend because they are the location of the Calvin–Benson–Bassham (CBB) cycle responsible for CO2 assimilation [1]. This process uses ATP and NADPH produced in the thylakoids by the primary phase of photosynthesis, and involves 11 enzymes that catalyse 13 reactions. It comprises three main stages: a carboxylation step performed by ribulose-1,5-bisphosphate carboxylase-oxygenase; a reduction of phosphoglycerate performed by phosphoglycerate kinase (PGK) and NADP-dependent glyceraldehyde-3-phosphate dehydrogenase at the expense of ATP and NADPH; and a regeneration step that converts triose-phosphate into ribulose-1,5-bisphosphate involving a series of enzymes (from triose phosphate isomerase and fructose-1,6-bisphosphatase (FBPase) to phosphoribulokinase (PRK)).
The complexity and central role of the CBB cycle within a wider metabolic network, plus the variations in the supply of light, CO2 and other resources, require the enzymes involved to be finely regulated. In the well-studied higher plants and green algae, the CBB cycle is active in the light and inactive in the dark and the main known regulatory mechanisms involve pH, Mg2+, redox-state, metabolites and protein–protein interactions [2,3]. One of the best known protein–protein interactions is transitory, linked to light-regulation and involves thioredoxin-mediated reversible reduction/oxidation of disulfide bonds/sulfhydryl group of cysteine residues [4]. In the light, thioredoxins (TRXs) are reduced by electrons from photosystem I and, in turn, reduce their target enzymes, while in the dark they are oxidized and then oxidize their targets. Diatoms have many different thioredoxins each encoded by a specific gene and located in different compartments, including the chloroplast; most contain the regulatory cysteine residues in the conserved motif, WCGPC [5]. Glutathionylation is another mechanism of redox regulation that can regulate CBB enzymes from photosynthetic organisms and some targets have been proposed [6], but little is known in diatoms and this is therefore not covered here. Another type of regulation found in higher plants, some algae and some cyanobacteria, involves the formation of multi-enzyme complexes that modify the regulatory and kinetic properties of the component enzymes [7,8]. Lastly, carbon fixation is also regulated at the transcriptional level [9,10].
Diatoms (Bacillariophyceae) comprise 30 000–100 000 species [11], are ecologically widespread [12] and, despite having evolved relatively recently, around 250 million years ago [13], are major players in global biogeochemical cycles [14]. They originated via a secondary endosymbiosis and their chloroplasts derive from a red algal ancestor. Their pigments are chlorophyll a, c and fucoxanthin, rather than red algal chlorophyll a and phycoerythrin or green algal chlorophyll a and b, and photosynthesis products are stored as chrysolaminarin and lipid rather than red algal floridean starch or green algal starch. However, they have an enigmatic evolutionary history and also contain green algal, bacterial and animal-like genes [15–17]. Diatoms have a number of ‘variations’ on the original Plantae (Archaeplastida) theme including: the possession of a functioning urea cycle [18] and a Entner–Doudoroff glycolytic pathway [19], and lack the oxidative pentose phosphate (OPP) pathway in the chloroplast [20,21]. Since OPP produces ribose-5-phosphate used in the synthesis of nucleotides, its absence in the chloroplast requires pyrimidine and purine nucleotides that were synthesized in the cytosol to be transported into the chloroplast [22,23]. Because of all these differences, one might expect the CBB cycle in diatoms to be regulated differently from those in the Plantae.
2. Ribulose-1,5-bisphosphate carboxylase-oxygenase and its activase
The crux of the CBB cycle is the rate-limiting step catalysed by ribulose-1,5-bisphosphate carboxylase-oxygenase (Rubisco), the most abundant protein on Earth [24,25]. However, it has been shown that the levels of Rubisco have little impact on the control of carbon fixation and therefore this enzyme does not necessarily constrain photosynthesis under normal conditions [26]. Rubisco catalyses the carboxylation of ribulose-1,5-bisphosphate (RuBP) by CO2 to give two molecules of 3-phosphoglycerate (PGA). The turnover number (kc) of Rubisco, around 5 s−1, is among the lowest for any enzyme and its catalytic efficiency (kc/Km) and affinity for CO2 are low [27,28]. Furthermore, depending on the relative concentration of CO2 and O2 at the active site, and the specificity of the enzyme, Rubisco can catalyse an oxygenation reaction that reduces the rate of net photosynthesis and produces PGA and phosphoglycolate [29] that is metabolized through the photorespiratory pathway [30].
Crystal structures [31,32] show that the active site of Rubisco is formed by the interaction of the L subunits, in a head-to-tail arrangement with the C-terminus of one monomer next to the N-terminus of a second monomer to form two active sites per dimer; however, no structural information is available for Rubisco from diatoms. The CO2/O2 specificity factor Ω, (also called τ) is variable and for Rubisco from land plants is in the range 80–100 [33]. The specificity seems to have increased during evolution to compensate for the gradual atmosphere decrease in CO2, and increase in O2 [34]. As a result, the more recently evolved higher plants and diatoms, though having different forms of Rubisco (forms 1B and 1D respectively), have, in general, higher values of Ω (around 110) than cyanobacteria (around 47) and other algae (66 for green algae) [35,36] implying positive selection of diatom and land plant Rubisco characteristics [37]. By contrast, the Km for CO2 of diatom Rubisco is highly variable (from 23 to 60 µM [38]) and greater than that found in land plants (around 10 µM [38]).
In aquatic systems, CO2 diffusion is low and the CO2 concentration is often below the relatively high Km[CO2] of diatom Rubisco; therefore, a CO2-concentrating mechanism (CCM) is beneficial [38–41]. Different forms of CCMs exist involving bicarbonate transport, and, contentiously, C4 metabolism [42]. In both energy-requiring processes, intra- or extracellular carbonic anhydrases (CA) are involved to maintain chemical equilibrium between CO2 and bicarbonate. In diatoms, the location and number of CAs are variable [43] and CAs are redox-regulated by TRX in Phaeodactylum tricornutum [44]. To our knowledge, this has not been reported in other photoautotrophs and whether or not it occurs in other diatoms is currently unknown.
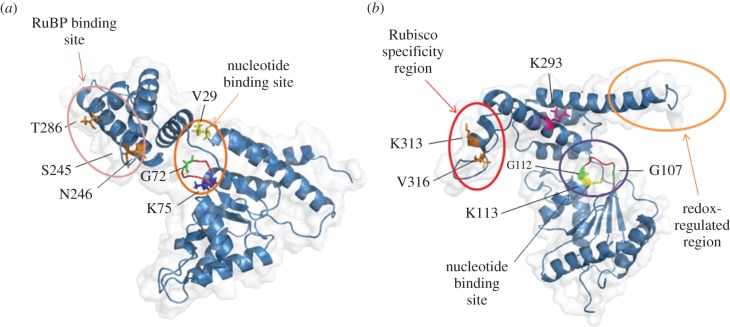
Regulation of Rubisco matches the CO2 fixation rate to the photosynthetic electron transport rate, helping to adjust the concentration of chloroplast metabolites to the demand by photosynthesis [45]. All bona fide Rubisco must first be activated or carbamylated at a lysine residue (e.g. Lys-201 of the spinach enzyme [46]) by a non-substrate CO2. Carbamate formation is favoured by alkaline pH and high concentrations of Mg2+ in the stroma. Not only Rubisco is regulated by these changes in Plantae, but also PRK, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the two phosphatases from the CBB cycle [3]. In plants, Rubisco has been extensively studied and can form multi-enzyme complexes with other CBB enzymes (for review see [3]). Rubisco is also regulated by Rubisco activase (RCA) [47]. RCA binds to inactive Rubisco and upon ATP hydrolysis, changes the conformation of the enzyme, thereby generating a highly active form of Rubisco. RCA thus works along with pH and Mg2+ to match the activity of Rubisco to the supply of resources. RCA is a member of the AAA+ (ATPase associated with diverse cellular activities) protein superfamily [47,48]. RCA can be redox regulated at its C-terminus, which contains two cysteine residues (Cys392 and Cys411) that form a disulfide bridge under oxidized conditions, producing a conformational change that blocks ATP interaction with RCA [49]. Surprisingly, no homologue of RCA is known for red type Rubisco. Recently however, a crystal structure of a protein that plays the activase role for the red type Rubisco, CbbX (Calvin–Benson–Bassham protein X), was described in Rhodobacter sphaeroides [50]. CbbX also shows the typical AAA+ hexameric conformation and has functional analogy with RCA despite their low sequence homology (18%). Using BLAST software we found CbbX homologues in all the stramenopiles (more than 100) that we could check, and also showed that the CbbX gene is encoded in the chloroplast genome, unlike the RCA gene which is located in the nuclear genome. Using CbbX red-type Rubisco from R. sphaeroides (3ZUH.pdb) as a template, we found that the structures of CbbX in Thalassiosira pseudonana, P. tricornutum and Asterionella formosa were similar (data not shown). Some slight differences were found between Arabidopsis thaliana RCA and P. tricornutum CbbX that are both regulated through their C-terminus. Phaeodactylum tricornutum. CbbX (figure 1a), unlike RCA, does not have cysteine residues that could provide redox sensitivity; however, it is regulated upon RuBP binding at its C-terminus [50]. The presence of six amino acid residues within a long flexible C-terminal tail found in red type Rubisco from R. sphaeroides, and also present in diatoms (figure 2a), seems to be necessary for fully activating the CbbX ATPase activity [50]. We hypothesize that the allosteric regulation of the CbbX by RuBP found in R. sphaeroides will also be present in the diatoms and will regulate Rubisco.
Figure 1.
Comparison of CbbX and RCA. CbbX monomer from P. tricornutum (a) is compared with RCA from A. thaliana (PDB: 4W5 W; (b)). Important residues are identified and highlighted. Residues are numbered according to P. tricornutum and A. thaliana respective sequences. The ‘P-loop’ (Walker-A) is also shown (in red) in both models. In CbbX, key amino acids for RuBP and nucleotide binding sites were predicted according to the R. sphaeroides model [50,51]. Residues K313 and V316 in RCA are involved in RCA-Rubisco specificity [49,52]. Residues involved in RCA ATP binding site—K113 [52,53], G112, G107 [54]—and ATP hydrolysis—K293 [52]—are shown. A redox-regulated region (see [49]) is also identified, although part of the C-terminus is missing in the representation. Phaeodactylum tricornutum CbbX was modelled using SWISS MODEL webtool and R. sphaeroides CbbX crystal structure (PDB: 3SYL) as template. Ribbon representations were produced with PyMol software.
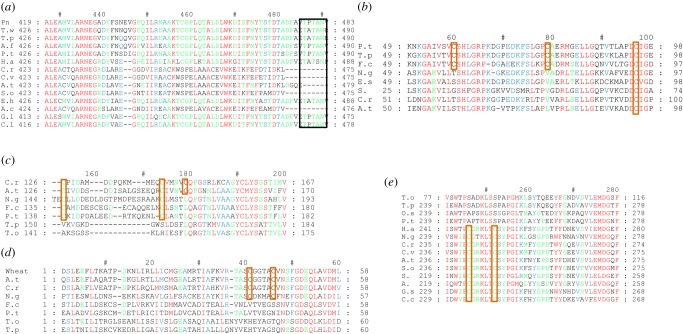
Figure 2.
Alignment of Calvin cycle enzymes. (a) Rubisco C-terminal region alignment. Black rectangle shows C-terminal extension; (b) PGK; (c) FBPase; (d) SBPase; (e) PRK. Species used are the following: Green algae: C.r: Chlamydomonas reinhardtii, V.c: Volvox carteri, C.v: Chlorella vulgaris; Diatoms: Pn: Pseudo-nitzchia sp., T.w: Thalassiosira weisflogii, T.p: Thalassiosira pseudonana, T.o: Thalassiosira oceanica, P.t: Phaeodactylum tricornutum, A.f: Asterionella formosa, O.s: Odontella sinensis; F.c: Fragilariopsis cylindrus; Coccolithophores: E.h: Emiliania huxleyi; Other stramenopiles: H.a: Heterosigma akashiwo, N.g: Nannochloropsis gaditana; Rhodophytes: G.l: Gloiocladia laciniata, C.l: Chondria littoralis; Cyanobacteria: S.: Synechococcus sp. PCC 7336, S. sp: Synechocystis sp., A.c: Anabaena cylindrica, A.: Anabaena sp.; Land plants: A.t: Arabidopsis thaliana, S.o: Spinacia oleracea, B.n: Brassica napus, P.m: Prunus mume, S.l: Solanum lycopersicum. Redox-regulated cysteine residues are highlighted by an orange rectangle. Alignments were performed with ClustalW, through MEGA6 software, and processed by GenDoc software.
3. Phosphoglycerate kinase
PGK is involved in several pathways, such as glycolysis and gluconeogenesis, and phosphorylates 3-phosphoglycerate (PGA) with ATP, producing 1,3-biphosphoglycerate (BPGA). Different isoforms are located in different compartments [55]. In higher plants, PGK is thought to have a cyanobacterial origin which, after subsequent gene duplication, gave rise to the chloroplastic and cytosolic enzymes [56,57]. Green algae also have a cyanobacterial-derived PGK. Previously, it was thought that green algae only had a chloroplastic form of PGK [58], but subsequently many cytosolic PGK sequences have been found in green algae [59,60]. In diatoms, the number of PGK isoenzymes and their subcellular location is also variable [22]. A comparative analysis of the sequenced genomes of the diatoms, P. tricornutum, T. pseudonana and Fragilariopsis cylindrus [61] showed that PGK isoforms in the cytosol, mitochondria and chloroplast are different, and that more than one isoform can occur within the same subcellular compartment. The variation in PGK isoforms among diatom species is greater in the cytosol than in the mitochondria and chloroplasts, indicating that cytosolic PGK genes were less conserved during diatom diversification. In comparison, diatom chloroplastic PGK is more conserved and more closely related to the red algae from which they derive [62].
Chloroplastic PGK can be redox-regulated by TRXs and specifically the f isoform [63]. In Chlamydomonas reinhardtii, and in the cyanobacterium Synechocystis sp. PCC6803, TRX-mediated regulation of PGK involves the formation/dissociation of a disulfide bond between Cys265 and Cys99 [63], and Cys352 and Cys378 (numbering from diatom PGK [58,64]), respectively. However, PGK might not be regulated in all green algae, land plants or cyanobacteria because some species lack critical cysteine residues. In contrast to other CBB enzymes, PGK is regulated in at least some diatoms. In P. tricornutum, chloroplastic PGK is a target of TRX and can be inactivated by oxidation [65]. This might be a consequence of sulfenic acid formation on the Cys80 (–SOH) and subsequent disulfide bond formation between Cys61 and Cys99. These three cysteine residues are also present in T. pseudonana and F. cylindrus, suggesting a common mechanism among diatoms, but they are absent in other stramenopiles (figure 2b).
4. Glyceraldehyde 3-phosphate dehydrogenase and CP12
GAPDH exists as three main forms in higher plants and algae, two cytosolic forms involved in gluconeogenesis and glycolysis, and a chloroplastic form involved in the CBB cycle. GAPDH isolated from chloroplasts can use either NAD(H) or NADP(H) to catalyse the reversible reduction and dephosphorylation of 1,3-bisphosphoglycerate (BPGA) to produce glyceraldehyde-3-phosphate and inorganic phosphate. In vascular plants, GAPDH exists either as a heterotetramer of two A subunits and two B subunits (A2B2), or as a homotetramer of four A subunits (A4) [66]. In red and green algae, only the A subunit exists. The A and B subunits are very similar, except that the B subunit has a C-terminal extension of 30 amino acid residues that contains two cysteine residues homologous to the CP12 C-terminus [67]. Diatoms, like most stramenopiles, have a chloroplastic GAPDH comprising four C1 subunits, similar to the cytosolic form in all studied species [68,69] which, like the A subunit of GAPDH, does not have a CP12-homologous region at its C-terminus.
CP12 is a chloroplastic intrinsically disordered protein, involved in GAPDH redox regulation and in the formation of a GAPDH-CP12-PRK complex that has been found in land plants [70], green and red algae [71–73] and the cyanobacterium Synechococcus elongatus [74]. The complex forms in oxidizing conditions (i.e. during the night), with inactive enzymes, and dissociates in reducing conditions (i.e. during the day) through TRX reduction, releasing active enzymes [70,75]. CP12 sequences usually contain four cysteine residues. Two of them (Cys23 and Cys31) are important for the association of CP12 with PRK, whereas the two other cysteine residues (Cys 66 and Cys75) are important for GAPDH binding [72,76,77]. A highly conserved domain (WXXVEEXXXXXH) is also present, is located in the middle of CP12 sequence and is involved in the formation of the CP12-PRK complex, but not the CP12-GAPDH complex [78].The CP12 sequences in diatoms differ from those from land plants, cyanobacteria and green algae to the extent that it has been suggested that CP12 is absent in diatoms [17]. A protein (ID: XP_002288136.1) found in T. pseudonana has 35 per cent identity with C. reinhardtii CP12 and has the (VAWDXVEELXAAXSHK) sequence and the two N-terminal cysteine residues. In P. tricornutum, a CP12-like protein is present that has a 38% percent identity with the green algal CP12 and a TSPEARVAWDAVEEM sequence, but lacks the four cysteine residues. Interestingly, in diatoms, though the presence of the complex mentioned above has not been shown, another complex involving GAPDH, a CP12-like protein and ferredoxin-NADP reductase (FNR) has been described in the freshwater diatom A. formosa [79]. In this diatom, the redox regulation of GAPDH was dependent on the presence of the CP12-like protein, as when GAPDH was dissociated from this protein, the redox regulation was absent. Another regulation of GAPDH by NADP(H) occurs when GAPDH interacts with the CP12-like protein in the presence of FNR [79]. GAPDH regulation in diatoms, however, seems to vary from one species to another [80] and more work is required on this enzyme as well as on the CP12-like proteins in these organisms.
5. Fructose-1,6-bisphosphatase and sedoheptulose-1,7-bisphosphatase
FBPase and sedoheptulose-1,7-bisphosphatase (SBPase) irreversibly catalyse the dephosphorylation of fructose-1,6-bisphosphate (FBP) and sedoheptulose-1,7-bisphosphate (SBP) producing fructose-6-phosphate and sedoheptulose-7-phosphate, respectively [81,82]. SBPase is unique to the CBB cycle, while FBPase is involved in several pathways. Chloroplastic FBPases of red and green algae, and plants are not of cyanobacterial origin but instead are believed to have evolved from cytosolic forms through gene duplication [83,84]. Similarly, because cyanobacteria lack SBPase and instead use a FBPase that can hydrolyse both FBP and SBP, it is likely that the plastid SBPase did not originate from cyanobacteria [85]. While FBPase involved in gluconeogenesis is regulated by fructose-2,6-bisphosphate and AMP, similarly to the mammalian enzyme [86], the chloroplastic FBPase, like SBPase, is regulated by light via TRXs [87]. Both phosphatases are also regulated by light-induced changes in stromal Mg2+ and pH [88]. Chloroplastic FBPases in Plantae bear three conserved cysteine residues at positions 155, 174 and 179 (numbered from spinach enzyme) that are responsible for redox regulation. These residues are absent in cytosolic enzymes that are consequently redox-insensitive [89]. In diatoms, the regulatory cysteine residues, Cys153, Cys174 and Cys179 (numbering from green algae and higher plants) are absent in the chloroplastic FBPases from Thalassiosira oceanica and T. pseudonana, while only Cys153 and Cys174 are present in P. tricornutum, F. cylindrus and in the eustigmatophyte Nannochloropsis gaditana (figure 2c). For Odontella sinensis, it has been mentioned that FBPase and SBPase are redox-regulated, but no data were presented [90] and unfortunately no sequences are available for these enzymes from this species. Whether or not FBPase is redox-regulated in diatoms requires further investigation.
SBPases from Plantae bear two regulatory cysteine residues, Cys41 and Cys46 (numbering from green algae and higher plants), and are highly redox-regulated. Based on this high regulation, on antisense RNA techniques, and on modelling studies in land plants, it seems that SBPase, even more than FBPase, plays an important role in the control of carbon flux through the CBB cycle in land plants [26]. The SBPases from diatoms lack these regulatory cysteine residues (figure 2d), suggesting that they are unlikely to be redox-regulated, and they are not regulated at the transcript level [91], suggesting that this enzyme plays a less strategic role in diatoms than in Plantae.
6. Phosphoribulokinase
PRK is unique to the CBB cycle, and catalyses the ATP-Mg2+-dependent phosphorylation of ribulose-5-phosphate (Ru5P), thus regenerating RuBP, the Rubisco substrate. PRKs from different organisms differ in their catalytic and regulatory properties and their oligomerization state. In anoxygenic photosynthetic bacteria such as R. sphaeroides, PRK is octameric, inhibited by AMP, allosterically activated by NADH [92] and its structure has been solved at a resolution of 2.6 Å [93]. In some well-studied oxygenic phototrophs, PRK is dimeric and regulated by redox-sensitive cysteine residues and/or inhibitory GAPDH-CP12-PRK complex formation [73]. The activity of PRK from the diatom O. sinensis is not affected by light or dark treatment [90]. Moreover, PRK from the freshwater diatom A. formosa is insensitive to a reducing agent, dithiothreitol, that mimicks in vitro the action of TRX in vivo [94]. It is currently unknown if the absence of the OPP pathway from the chloroplast is functionally linked (how and why) to the lack of redox-regulation of PRK [20,90]. A survey of PRK activities in species from many phylogenetic groups showed that not only the presence of the two regulatory cysteine residues, but also the distance between them, determined whether or not the enzyme was redox-regulated. For example, PRKs from the Chlorophyta all have 38 residues between the two cysteine residues at positions 16 and 55 (numbering from the spinach and green algal enzymes), and enzymes from this group are redox-regulated [80]. In contrast, in diatoms the distance between the two regulatory cysteine residues is five amino acid residues longer than in land plants and green algae, and apparently this insertion is sufficient to prevent PRK redox-regulation.
In the green alga C. reinhardtii, the arginine residue at position 64 in PRK is important both for the binding of Ru5P [95] and for the formation of the supramolecular complex with GAPDH [96]. Recently, it has been shown that the formation of a disulfide bridge between Cys243 and Cys249 in C. reinhardtii is essential for the formation of the ternary complex involving PRK-CP12-GAPDH [97]. These cysteine residues are highly conserved and located at the same position among diverse photosynthetic organisms such as in A. thaliana and S. elongatus where the complex is also present, but are absent in PRK from diatoms (figure 2e). This suggests that diatom PRK is not able to form a complex with CP12 and GAPDH, and hitherto this complex has not been found in any diatoms. To conclude, PRK in diatoms does not seem to be regulated by redox process or by protein–protein interaction upon light/dark transition but could be regulated at the transcript level as shown in P. tricornutum [91] and possibly furthermore by other mechanisms yet to be discovered.
7. Transcriptional regulation of Calvin–Benson–Bassham enzymes
Transcriptional regulation of CBB enzymes could be an important adaptive response of photosynthetic organisms to environmental variation [98], but relatively little is known about this. In land plants, CBB genes are targets for gene regulation under light variation [10] and abiotic stresses [99]. Moreover, upstream regulatory motifs for 12 CBB genes [10] and transcription factors (TF) have been identified [98,100], most of them belonging to the bZIP family. However, in diatoms, we were not able to find any homologues of the TFs [100] although some proteins containing the bZIP motif were present in their genome (data not shown). In C. reinhardtii, a TF (CIA5) involved in CCM-related gene expression, is thought to be a negative regulator of some CBB genes, but the mechanism is not yet clear [101]. However, this TF seems to be exclusive to green algae because no homologues were found in other organisms.
In A. thaliana, the expression of the genes for CP12s, PRK and GAPDH is tissue-specific [102] and, more interestingly, is coordinated, consistent with the formation of a ternary complex between the three proteins [103]. In P. tricornutum, the genes encoding PRK, and the chloroplastic but not the cytosolic GAPDH, are regulated by light/dark transition [91]. This suggests that the mechanisms of gene expression of the two GAPDH homologues are probably different. In C. reinhardtii, the SBPase gene csbp is light-regulated by its 1.4 kb upstream region [104]. In contrast, in P. tricornutum, as mentioned above, both the SBPase gene and protein are unaffected by light or dark, a characteristic that might be shared with other diatoms. However, triose phosphate isomerase, FBPase and fructose-bisphosphate aldolase are highly upregulated after 24–48 h under high light in this species [105].
Further evidence that CBB enzymes from diatoms are regulated at the gene level derives from nutrient starvation experiments. A comparison of the transcriptomes of different diatoms showed that CBB genes were generally downregulated under nitrate deprivation [106]. Similarly, in N-deprived P. tricornutum, most CBB genes were downregulated while OPP pathway genes were upregulated [107]. In silicon-starved T. pseudonana, rbcR (or ycf30), a gene encoding for a Rubisco transcriptional regulator, was downregulated while lipid biosynthesis genes were upregulated [108]. In the red alga, Cyanidioschyzon merolae, rbcR upregulates the rbcLS-cbbx operon that contains the rbcLS gene, encoding for the Rubisco large and small subunit, during dark-to-light transition [109]. This operon is present in red algae, raphidophytes and α-proteobacteria, while the rbcLS-cbbx operon is absent in diatoms and coccolithophores. Therefore, cbbx and rbcLS are not under the control of the same promoter. However, all organisms, including diatoms, have rbcR and rbcLS that could be controlled by this regulator. In P. tricornutum, the rbcLS gene is highly upregulated during the first 6 h of a 12 h photoperiod, although its expression was not correlated with CO2 fixation [110]. Similarly, a lag between the maximum transcript level (at 09.00) and the maximal level of Rubisco (at 15.00) has been observed in T. pseudonana [111].
8. Conclusion
Regulation of the CBB cycle in diatoms is understudied relative to their ecological importance and seems to be very different from the Plantae. Given that there appears to be a large range of variability of CBB properties and regulation among diatoms, consistent with the large amount of genetic variation within the group [112], more species clearly need to be investigated.
Data accessibility
This article has no data.
Authors' contributions
All authors contribute to the writing and discussion of this review and E.J. also prepared both the figures.
Competing interests
We have no competing interests.
Funding
E.J.'s studentship is supported by Comisión Nacional de Investigación Científica y Tecnológica (CONiCyT), Chile, and Romain Clément's studentship was supported by the Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT). B.G.'s group is supported by Centre National de la Recherche Scientifique, Aix-Marseille Université, A*midex project (no. ANR-11-IDEX-0001-02), Agence Nationale de la Recherche (Signaux-BioNRJ, ANR-15-CE05-0021-03), the Region PACA. S.C.M.'s work is supported by the UK Natural Environment Research Council and a visiting scholarship from Aix-Marseille Université.
References
- 1.Buchanan BB. 2016. The carbon (formerly dark) reactions of photosynthesis. Photosynth. Res. 128, 215–217. ( 10.1007/s11120-015-0212-z) [DOI] [PubMed] [Google Scholar]
- 2.Michelet L, et al. 2013. Redox regulation of the Calvin-Benson cycle: something old, something new. Front. Plant Sci. 4, 470 ( 10.3389/fpls.2013.00470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gontero B, Avilan L, Lebreton S. 2006. Control of carbon fixation in chloroplasts. Annu. Plant Rev. 22, 187–218. [Google Scholar]
- 4.Buchanan BB. 1980. Role of light in the regulation of chloroplast enzymes. Annu. Rev. Plant Physiol. 31, 341–374. ( 10.1146/annurev.pp.31.060180.002013) [DOI] [Google Scholar]
- 5.Weber T, Gruber A, Kroth PG. 2009. The presence and localization of thioredoxins in diatoms, unicellular algae of secondary endosymbiotic origin. Mol. Plant 2, 468–477. ( 10.1093/mp/ssp010) [DOI] [PubMed] [Google Scholar]
- 6.Rouhier N, Lemaire SD, Jacquot JP. 2008. The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166. ( 10.1146/annurev.arplant.59.032607.092811) [DOI] [PubMed] [Google Scholar]
- 7.Lebreton S, Graciet E, Gontero B. 2003. Modulation, via protein-protein interactions, of glyceraldehyde-3-phosphate dehydrogenase activity through redox phosphoribulokinase regulation. J. Biol. Chem. 278, 12 078–12 084. ( 10.1074/jbc.M213096200) [DOI] [PubMed] [Google Scholar]
- 8.Lebreton S, Gontero B. 1999. Memory and imprinting in multienzyme complexes. Evidence for information transfer from glyceraldehyde-3-phosphate dehydrogenase to phosphoribulokinase under reduced state in Chlamydomonas reinhardtii. J. Biol. Chem. 274, 20 879–20 884. ( 10.1074/jbc.274.30.20879) [DOI] [PubMed] [Google Scholar]
- 9.Fey V, Wagner R, Brautigam K, Pfannschmidt T. 2005. Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 56, 1491–1498. ( 10.1093/jxb/eri180) [DOI] [PubMed] [Google Scholar]
- 10.Sun N, Ma L, Pan D, Zhao H, Deng XW. 2003. Evaluation of light regulatory potential of Calvin cycle steps based on large-scale gene expression profiling data. Plant Mol. Biol. 53, 467–478. ( 10.1023/B:PLAN.0000019071.12878.9e) [DOI] [PubMed] [Google Scholar]
- 11.Mann DG, Vanormelingen P. 2013. An inordinate fondness? The number, distributions, and origins of diatom species. J. Eukaryot. Microbiol. 60, 414–420. ( 10.1111/jeu.12047) [DOI] [PubMed] [Google Scholar]
- 12.Armbrust EV, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. ( 10.1126/science.1101156) [DOI] [PubMed] [Google Scholar]
- 13.Medlin LK. 2016. Evolution of the diatoms: major steps in their evolution and a review of the supporting molecular and morphological evidence. Phycologia 55, 79–103. ( 10.2216/15-105.1) [DOI] [Google Scholar]
- 14.Granum E, Raven JA, Leegood RC. 2005. How do marine diatoms fix 10 billion tonnes of inorganic carbon per year? Can. J. Bot. 8, 898–908. ( 10.1139/b05-077) [DOI] [Google Scholar]
- 15.Deschamps P, Moreira D. 2012. Reevaluating the green contribution to diatom genomes. Gen. Biol. Evol. 4, 795–800. ( 10.1093/gbe/evs053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. 2009. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726. ( 10.1126/science.1172983) [DOI] [PubMed] [Google Scholar]
- 17.Wilhelm C, et al. 2006. The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae. Protist 157, 91–124. ( 10.1016/j.protis.2006.02.003) [DOI] [PubMed] [Google Scholar]
- 18.Allen AE, et al. 2011. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207. ( 10.1038/nature10074) [DOI] [PubMed] [Google Scholar]
- 19.Fabris M, Matthijs M, Rombauts S, Vyverman W, Goossens A, Baart GJ. 2012. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 70, 1004–1014. ( 10.1111/j.1365-313X.2012.04941.x) [DOI] [PubMed] [Google Scholar]
- 20.Gruber A, Weber T, Bartulos CR, Vugrinec S, Kroth PG. 2009. Intracellular distribution of the reductive and oxidative pentose phosphate pathways in two diatoms. J. Basic Microbiol. 49, 58–72. ( 10.1002/jobm.200800339) [DOI] [PubMed] [Google Scholar]
- 21.Kroth PG. 2015. The biodiversity of carbon assimilation. J. Plant Physiol. 172, 76–81. ( 10.1016/j.jplph.2014.07.021) [DOI] [PubMed] [Google Scholar]
- 22.Kroth PG, et al. 2008. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3, e1426 ( 10.1371/journal.pone.0001426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ast M, Gruber A, Schmitz-Esser S, Neuhaus HE, Kroth PG, Horn M, Haferkamp I. 2009. Diatom plastids depend on nucleotide import from the cytosol. Proc. Natl Acad. Sci. USA 106, 3621–3626. ( 10.1073/pnas.0808862106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raven JA. 2013. Rubisco: still the most abundant protein of Earth? New phytol. 198, 1–3. ( 10.1111/nph.12197) [DOI] [PubMed] [Google Scholar]
- 25.Ellis RJ. 1979. The most abundant protein in the world. Trends Biochem. Sci. 4, 241–244. ( 10.1016/0968-0004(79)90212-3) [DOI] [Google Scholar]
- 26.Raines CA. 2003. The Calvin cycle revisited. Photosynth. Res. 75, 1–10. ( 10.1023/A:1022421515027) [DOI] [PubMed] [Google Scholar]
- 27.Cleland WW, Andrews TJ, Gutteridge S, Hartman FC, Lorimer GH. 1998. Mechanism of Rubisco: the carbamate as general base. Chem. Rev. 98, 549–562. ( 10.1021/cr970010r) [DOI] [PubMed] [Google Scholar]
- 28.Tabita FR. 1999. Microbial ribulose bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60, 1–28. ( 10.1023/A:1006211417981) [DOI] [Google Scholar]
- 29.Bowes G, Ogren WL. 1972. Oxygen inhibition and other properties of soybean ribulose 1,5-diphosphate carboxylase. J. Biol. Chem. 247, 2171–2176. [PubMed] [Google Scholar]
- 30.Moroney JV, Jungnick N, Dimario RJ, Longstreth DJ. 2013. Photorespiration and carbon concentrating mechanisms: two adaptations to high O2, low CO2 conditions. Photosynth. Res. 117, 121–131. ( 10.1007/s11120-013-9865-7) [DOI] [PubMed] [Google Scholar]
- 31.Schneider G, Knight S, Andersson I, Branden CI, Lindqvist Y, Lundqvist T. 1990. Comparison of the crystal structures of L2 and L8S8 Rubisco suggests a functional role for the small subunit. EMBO J. 9, 2045–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I. 2001. First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J. Biol. Chem. 276, 48 159–48 164. ( 10.1074/jbc.M107765200) [DOI] [PubMed] [Google Scholar]
- 33.Spreitzer RJ. 1999. Questions about the complexity of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase. Photosynth. Res. 60, 29–42. ( 10.1023/A:1006240202051) [DOI] [Google Scholar]
- 34.Tortell PD. 2000. Evolutionary and ecological perspectives on carbon acquisition in phytoplankton. Limnol. Oceanogr. 45, 744–750. ( 10.4319/lo.2000.45.3.0744) [DOI] [Google Scholar]
- 35.Galmés J, Flexas J, Keys AJ, Cifre J, Mitchell RAC, Madgwick PJ, Haslam RP, Medrano H, Parry MA. J. 2005. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ. 28, 571–579. ( 10.1111/j.1365-3040.2005.01300.x) [DOI] [Google Scholar]
- 36.Galmés J, Kapralov MV, Andralojc PJ, Conesa MÀ, Keys AJ, Parry MA. J, Flexas J. 2014. Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant Cell Environ. 37, 1989–2001. ( 10.1111/pce.12335) [DOI] [PubMed] [Google Scholar]
- 37.Young JN, Rickaby RE, Kapitonov VV, Filatov DA. 2012. Adaptative signals in algal Rubisco reveal a history of ancient atmospheric carbon dioxide. Phil. Trans. R. Soc. B 367, 483–492. ( 10.1098/rstb.2011.0145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young JN, Heureux AM, Sharwood RE, Rickaby RE, Morel FM, Whitney SM. 2016. Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J. Exp. Bot. 67, 3445–3456. ( 10.1093/jxb/erw163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopkinson BM, Dupont CL, Allen AE, Morela FMM. 2011. Efficiency of the CO2-concentrating mechanism of diatoms. Proc. Natl Acad. Sci. USA 108, 3830–3837. ( 10.1073/pnas.1018062108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giordano M, Beardall J, Raven JA. 2005. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56, 99–131. ( 10.1146/annurev.arplant.56.032604.144052) [DOI] [PubMed] [Google Scholar]
- 41.Matsuda Y, Hopkinson BM, Nakajima K, Dupont CL, Tsuji Y. 2017. Mechanisms of carbon dioxide acquisition and CO2 sensing in marine diatoms: a gateway to carbon metabolism. Phil. Trans. R. Soc. B 372, 20160403 ( 10.1098/rstb.2016.0403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clement R, Dimnet L, Maberly SC, Gontero B. 2016. The nature of the CO2-concentrating mechanisms in a marine diatom, Thalassiosira pseudonana. New Phytol. 209, 1417–1427. ( 10.1111/nph.13728) [DOI] [PubMed] [Google Scholar]
- 43.Samukawa M, Shen C, Hopkinson BM, Matsuda Y. 2014. Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth. Res. 121, 235–249. ( 10.1007/s11120-014-9967-x) [DOI] [PubMed] [Google Scholar]
- 44.Matsuda Y, Kroth PG. 2014. Carbon fixation in diatoms. In The structural basis of biological energy generation (ed. Hohmann-Marriott MF.), pp. 335–362. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 45.Perchorowicz JT, Raynes DA, Jensen RG. 1981. Light limitation of photosynthesis and activation of ribulose bisphosphate carboxylase in wheat seedlings. Proc. Natl Acad. Sci. USA 78, 2985–2989. ( 10.1073/pnas.78.5.2985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorimer GH, Badger MR, Andrews TJ. 1976. The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. Equilibria, kinetics, a suggested mechanism, and physiological implications. Biochemistry 15, 529–536. ( 10.1021/bi00648a012) [DOI] [PubMed] [Google Scholar]
- 47.Gontero B, Salvucci M. 2014. Regulation of photosynthetic carbon metabolism in aquatic and terrestrial organisms by Rubisco activase, redox-modulation and CP12. Aquat. Bot. 118, 14–23. ( 10.1016/j.aquabot.2014.05.011) [DOI] [Google Scholar]
- 48.Portis AR. 2001. The Rubisco activase-Rubisco system: an ATPase-dependent association that regulates photosynthesis. In Protein-protein interactions in plant biology (eds McManus MT, Laing WA, Allan AC), pp. 30–52. Sheffield, UK: Sheffield Academic Press. [Google Scholar]
- 49.Portis AR Jr, Li C, Wang D, Salvucci ME. 2008. Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59, 1597–1604. ( 10.1093/jxb/erm240) [DOI] [PubMed] [Google Scholar]
- 50.Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. 2011. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199. ( 10.1038/nature10568) [DOI] [PubMed] [Google Scholar]
- 51.Stotz M, Mueller-Cajar O, Ciniawsky S, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. 2011. Structure of green-type Rubisco activase from tobacco. Nat. Struct. Mol. Biol. 18, 1366–1370. ( 10.1038/nsmb.2171) [DOI] [PubMed] [Google Scholar]
- 52.Li C, Wang D, Portis AR Jr. 2006. Identification of critical arginine residues in the functioning of Rubisco activase. Arch. Biochem. Biophys. 450, 176–182. ( 10.1016/j.abb.2006.04.002) [DOI] [PubMed] [Google Scholar]
- 53.Salvucci ME, Rajagopalan K, Sievert G, Haley BE, Watt DS. 1993. Photoaffinity labeling of ribulose-1,5-bisphosphate carboxylase/oxygenase activase with ATP gamma-benzophenone. Identification of the ATP gamma-phosphate binding domain. J. Biol. Chem. 268, 14 239–14 244. [PubMed] [Google Scholar]
- 54.Kallis RP, Ewy RG, Portis AR Jr. 2000. Alteration of the adenine nucleotide response and increased Rubisco activation activity of Arabidopsis Rubisco activase by site-directed mutagenesis. Plant Physiol. 123, 1077–1086. ( 10.1104/pp.123.3.1077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin W, Scheibe R, Schnarrenberger C. 2000. The Calvin cycle and its regulation. In Photosynthesis: physiology and metabolism (eds Leegood RC, Sharkey TD, Caemmerer S), pp. 9–51. Dordrecht, The Netherlands: Kluwer Academic. [Google Scholar]
- 56.Brinkmann H, Martin W. 1996. Higher-plant chloroplast and cytosolic 3-phosphoglycerate kinases: a case of endosymbiotic gene replacement. Plant. Mol. Biol. 30, 65–75. ( 10.1007/BF00017803) [DOI] [PubMed] [Google Scholar]
- 57.Archibald JM, Keeling PJ. 2003. Comparative genomics. Plant genomes: cyanobacterial genes revealed. Heredity (Edinb) 90, 2–3. ( 10.1038/sj.hdy.6800204) [DOI] [PubMed] [Google Scholar]
- 58.Schnarrenberger C, Jacobshagen S, Muller B, Kruger I. 1990. Evolution of isozymes of sugar phosphate metabolism in green algae. Prog. Clin. Biol. Res. 344, 743–764. [PubMed] [Google Scholar]
- 59.Johnson X, Alric J. 2013. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot. Cell 12, 776–793. ( 10.1128/EC.00318-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin M, Turpin DH. 1993. Purification and characterization of two form of phosphoglycerate kinase from the green alga Selenastrum minutum. J. Phycol. 29, 777–786. ( 10.1111/j.0022-3646.1993.00777.x) [DOI] [Google Scholar]
- 61.Smith SR, Abbriano RM, Hildebrand M. 2012. Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Res. 1, 2–16. ( 10.1016/j.algal.2012.04.003) [DOI] [Google Scholar]
- 62.McFadden GI. 2001. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 37, 951–959. ( 10.1046/j.1529-8817.2001.01126.x) [DOI] [Google Scholar]
- 63.Morisse S, Michelet L, Bedhomme M, Marchand CH, Calvaresi M, Trost P, Fermani S, Zaffagnini M, Lemaire SD. 2014. Thioredoxin-dependent redox regulation of chloroplastic phosphoglycerate kinase from Chlamydomonas reinhardtii. J. Biol. Chem. 289, 30012–30024. ( 10.1074/jbc.M114.597997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsukamoto Y, Fukushima Y, Hara S, Hisabori T. 2013. Redox control of the activity of phosphoglycerate kinase in Synechocystis sp. PCC6803. Plant Cell Physiol. 54, 484–491. ( 10.1093/pcp/pct002) [DOI] [PubMed] [Google Scholar]
- 65.Bosco MB, Aleanzi MC, Iglesias AA. 2012. Plastidic phosphoglycerate kinase from Phaeodactylum tricornutum: on the critical role of cysteine residues for the enzyme function. Protist 163, 188–203. ( 10.1016/j.protis.2011.07.001) [DOI] [PubMed] [Google Scholar]
- 66.Cerff R. 1979. Quaternary structure of higher plant glyceraldehyde-3-phosphate dehydrogenases. Eur. J. Biochem. 94, 243–247. ( 10.1111/j.1432-1033.1979.tb12891.x) [DOI] [PubMed] [Google Scholar]
- 67.Pohlmeyer K, Paap BK, Soll J, Wedel N. 1996. CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol. Biol. 32, 969–978. ( 10.1007/BF00020493) [DOI] [PubMed] [Google Scholar]
- 68.Liaud MF, Lichtle C, Apt K, Martin W, Cerff R. 2000. Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 17, 213–223. ( 10.1093/oxfordjournals.molbev.a026301) [DOI] [PubMed] [Google Scholar]
- 69.Takishita K, Yamaguchi H, Maruyama T, Inagaki Y. 2009. A hypothesis for the evolution of nuclear-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase genes in ‘Chromalveolate’ members. PLoS ONE 4, e4737 ( 10.1371/journal.pone.0004737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howard TP, Metodiev M, Lloyd JC, Raines CA. 2008. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc. Natl Acad. Sci. USA 105, 4056–4061. ( 10.1073/pnas.0710518105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oesterhelt C, Klocke S, Holtgrefe S, Linke V, Weber AP, Scheibe R. 2007. Redox regulation of chloroplast enzymes in Galdieria sulphuraria in view of eukaryotic evolution. Plant Cell Physiol. 48, 1359–1373. ( 10.1093/pcp/pcm108) [DOI] [PubMed] [Google Scholar]
- 72.Groben R, Kaloudas D, Raines CA, Offmann B, Maberly SC, Gontero B. 2010. Comparative sequence analysis of CP12, a small protein involved in the formation of a Calvin cycle complex in photosynthetic organisms. Photosynth. Res. 103, 183–194. ( 10.1007/s11120-010-9542-z) [DOI] [PubMed] [Google Scholar]
- 73.Graciet E, Gans P, Wedel N, Lebreton S, Camadro JM, Gontero B. 2003. The small protein CP12: a protein linker for supramolecular complex assembly. Biochemistry 42, 8163–8170. ( 10.1021/bi034474x) [DOI] [PubMed] [Google Scholar]
- 74.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. 2005. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 42, 504–513. ( 10.1111/j.1365-313X.2005.02391.x) [DOI] [PubMed] [Google Scholar]
- 75.Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, Trost P. 2009. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2, 259–269. ( 10.1093/mp/ssn061) [DOI] [PubMed] [Google Scholar]
- 76.Erales J, Mekhalfi M, Woudstra M, Gontero B. 2011. Molecular mechanism of NADPH-glyceraldehyde-3-phosphate dehydrogenase regulation through the C-terminus of CP12 in Chlamydomonas reinhardtii. Biochemistry 50, 2881–2888. ( 10.1021/bi1020259) [DOI] [PubMed] [Google Scholar]
- 77.Wedel N, Soll J, Paap BK. 1997. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc. Natl Acad. Sci. USA 94, 10 479–10 484. ( 10.1073/pnas.94.19.10479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Avilan L, Puppo C, Erales J, Woudstra M, Lebrun R, Gontero B. 2012. CP12 residues involved in the formation and regulation of the glyceraldehyde-3-phosphate dehydrogenase CP12-phosphoribulokinase complex in Chlamydomonas reinhardtii. Mol. Biosyst. 8, 2994–3002. ( 10.1039/C2MB25244A.) [DOI] [PubMed] [Google Scholar]
- 79.Mekhalfi M, Puppo C, Avilan L, Lebrun R, Mansuelle P, Maberly SC, Gontero B. 2014. Glyceraldehyde-3-phosphate dehydrogenase is regulated by ferredoxin-NADP reductase in the diatom Asterionella formosa. New Phytol. 203, 414–423. ( 10.1111/nph.12820) [DOI] [PubMed] [Google Scholar]
- 80.Maberly SC, Courcelle C, Groben R, Gontero B. 2010. Phylogenetically-based variation in the regulation of the Calvin cycle enzymes, phosphoribulokinase and glyceraldehyde-3-phosphate dehydrogenase, in algae. J. Exp. Bot. 61 735–745. ( 10.1093/jxb/erp337) [DOI] [PubMed] [Google Scholar]
- 81.Breazale VD, Buchanan BB, Wolosiuk RA. 1978. Chloroplast sedoheptulose-1,7-bisphosphatase: evidence for regulation by the ferredoxin/thioredoxin system. Z. Naturforsch. 33c, 521–528. [Google Scholar]
- 82.Zimmermann G, Kelly GJ, Latzko E. 1976. Efficient purification and molecular properties of spinach chloroplast fructose 1,6-bisphosphatase. Eur. J. Biochem. 70, 361–367. ( 10.1111/j.1432-1033.1976.tb11025.x) [DOI] [PubMed] [Google Scholar]
- 83.Martin W, Mustafa AZ, Henze K, Schnarrenberger C. 1996. Higher-plant chloroplast and cytosolic fructose-1,6-bisphosphatase isoenzymes: origins via duplication rather than prokaryote-eukaryote divergence. Plant Mol. Biol. 32, 485–491. ( 10.1007/BF00019100) [DOI] [PubMed] [Google Scholar]
- 84.Gutle DD, et al. 2016. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl Acad. Sci. USA 113, 6779–6784. ( 10.1073/pnas.1606241113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rogers M, Keeling PJ. 2004. Lateral transfer and recompartmentalization of Calvin cycle enzymes of plants and algae. J. Mol. Evol. 58, 367–375. ( 10.1007/s00239-003-2558-7) [DOI] [PubMed] [Google Scholar]
- 86.Villeret V, Huang S, Zhang Y, Lipscomb WN. 1995. Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-d-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography. Biochemistry 34, 4307–4315. ( 10.1021/bi00013a020) [DOI] [PubMed] [Google Scholar]
- 87.Schurmann P, Jacquot JP. 2000. Plant thioredoxin systems revisited. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371–400. ( 10.1146/annurev.arplant.51.1.371) [DOI] [PubMed] [Google Scholar]
- 88.Baret P, Cadet F. 1997. Regulation of photosynthetic enzymes via redox systems. Biochem. Educ. 25, 24–26. ( 10.1016/s0307-4412(96)00133-1) [DOI] [Google Scholar]
- 89.Marcus F, Moberly L, Latshaw SP. 1988. Comparative amino acid sequence of fructose-1,6-bisphosphatases: identification of a region unique to the light-regulated chloroplast enzyme. Proc. Natl Acad. Sci. USA 85, 5379–5383. ( 10.1073/pnas.85.15.5379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Michels AK, Wedel N, Kroth PG. 2005. Diatom plastids possess a phosphoribulokinase with an altered regulation and no oxidative pentose phosphate pathway. Plant Physiol. 137, 911–920. ( 10.1104/pp.104.055285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM. 2013. Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 161, 1034–1048. ( 10.1104/pp.112.206177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Novak JS, Tabita FR. 1999. Molecular approaches to probe differential NADH activation of phosphoribulokinase isozymes from Rhodobacter sphaeroides. Arch. Biochem. Biophys. 363, 273–282. ( 10.1006/abbi.1998.1084) [DOI] [PubMed] [Google Scholar]
- 93.Harrison DHT, Runquist JA, Holub A, Miziorko HM. 1998. The crystal structure of phosphoribulokinase from Rhodobacter sphaeroides reveals a fold similar to that of adenylate kinase. Biochemistry 37, 5074–5085. ( 10.1021/bi972805y) [DOI] [PubMed] [Google Scholar]
- 94.Boggetto N, Gontero B, Maberly SC. 2007. Regulation of phosphoribulokinase and glyceraldehyde 3-phosphate dehydrogenase in a freshwater diatom, Asterionella formosa. J. Phycol. 43, 1227–1235. ( 10.1111/j.1529-8817.2007.00409.x) [DOI] [PubMed] [Google Scholar]
- 95.Roesler KR, Marcotte BL, Ogren WL. 1992. Functional importance of arginine 64 in Chlamydomonas reinhardtii phosphoribulokinase. Plant Physiol. 98, 1285–1289. ( 10.1104/pp.98.4.1285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avilan L, Gontero B, Lebreton S, Ricard J. 1997. Information transfer in multienzyme complexes-2. The role of Arg64 of Chlamydomonas reinhardtii phosphoribulokinase in the information transfer between glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase. Eur. J. Biochem. 250, 296–302. ( 10.1111/j.1432-1033.1997.0296a.x) [DOI] [PubMed] [Google Scholar]
- 97.Thieulin-Pardo G, Remy T, Lignon S, Lebrun R, Gontero B. 2015. Phosphoribulokinase from Chlamydomonas reinhardtii: a Benson-Calvin cycle enzyme enslaved to its cysteine residues. Mol. Biosyst. 11, 1134–1145. ( 10.1039/c5mb00035a) [DOI] [PubMed] [Google Scholar]
- 98.Saibo NJ, Lourenco T, Oliveira MM. 2009. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103, 609–623. ( 10.1093/aob/mcn227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nouri MZ, Moumeni A, Komatsu S. 2015. Abiotic stresses: insight into gene regulation and protein expression in photosynthetic pathways of plants. Int. J. Mol. Sci. 16, 20 392–20 416. ( 10.3390/ijms160920392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu X, Zheng G, Shan L, Meng G, Vingron M, Liu Q, Zhu XG. 2014. Reconstruction of gene regulatory network related to photosynthesis in Arabidopsis thaliana. Front. Plant Sci. 5, 273 ( 10.3389/fpls.2014.00273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fang W, Si Y, Douglass S, Casero D, Merchant SS, Pellegrini M, Ladunga I, Liu P, Spalding MH. 2012. Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24, 1876–1893. ( 10.1105/tpc.112.097949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh P, Kaloudas D, Raines CA. 2008. Expression analysis of the Arabidopsis CP12 gene family suggests novel roles for these proteins in roots and floral tissues. J. Exp. Bot. 59, 3975–3985. ( 10.1093/jxb/ern236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marri L, Sparla F, Pupillo P, Trost P. 2005. Co-ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. J. Exp. Bot. 56, 73–80. ( 10.1093/jxb/eri020) [DOI] [PubMed] [Google Scholar]
- 104.Hahn D, Kaltenbach C, Kück U. 1998. The Calvin cycle enzyme sedoheptulose-1,7-bisphosphatase is encoded by a light-regulated gene in Chlamydomonas reinhardtii. Plant Mol. Biol. 36, 929–934. ( 10.1023/a:1005911022601) [DOI] [PubMed] [Google Scholar]
- 105.Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM. 2009. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4, e7743 ( 10.1371/journal.pone.0007743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bender S, Durkin C, Berthiaume C, Morales R, Armbrust EV. 2014. Transcriptional responses of three model diatoms to nitrate limitation of growth. Front. Mar. Sci. 1, 3 ( 10.3389/fmars.2014.00003) [DOI] [Google Scholar]
- 107.Alipanah L, Rohloff J, Winge P, Bones AM, Brembu T. 2015. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 66, 6281–6296. ( 10.1093/jxb/erv340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith SR, Gle C, Abbriano RM, Traller JC, Davis A, Trentacoste E, Vernet M, Allen AE, Hildebrand M. 2016. Transcript level coordination of carbon pathways during silicon starvation-induced lipid accumulation in the diatom Thalassiosira pseudonana. New Phytol. 210, 890–904. ( 10.1111/nph.13843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Minoda A, Weber AP, Tanaka K, Miyagishima SY. 2010. Nucleus-independent control of the Rubisco operon by the plastid-encoded transcription factor Ycf30 in the red alga Cyanidioschyzon merolae. Plant Physiol. 154, 1532–1540. ( 10.1104/pp.110.163188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wawrik B, Paul JH, Tabita FR. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68, 3771–3779. ( 10.1128/aem.68.8.3771-3779.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Granum E, Roberts K, Raven JA, Leegood RC. 2009. Primary carbon and nitrogen metabolic gene expression in the diatom Thalassiosira pseudonana (Bacillariophyceae): diel periodicity and effects of inorganic carbon and nitrogen. J. Phycol. 45, 1083–1092. ( 10.1111/j.1529-8817.2009.00728.x) [DOI] [PubMed] [Google Scholar]
- 112.Bowler C, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. ( 10.1038/nature07410) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no data.