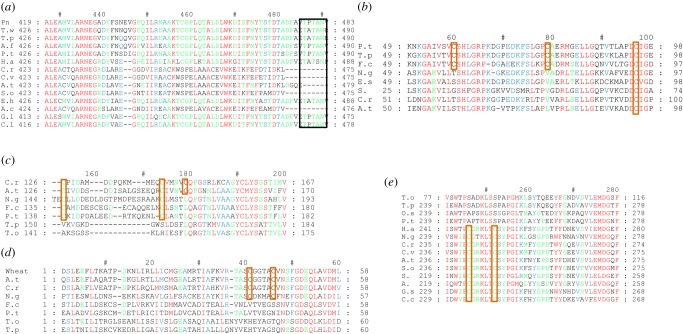
Figure 2.
Alignment of Calvin cycle enzymes. (a) Rubisco C-terminal region alignment. Black rectangle shows C-terminal extension; (b) PGK; (c) FBPase; (d) SBPase; (e) PRK. Species used are the following: Green algae: C.r: Chlamydomonas reinhardtii, V.c: Volvox carteri, C.v: Chlorella vulgaris; Diatoms: Pn: Pseudo-nitzchia sp., T.w: Thalassiosira weisflogii, T.p: Thalassiosira pseudonana, T.o: Thalassiosira oceanica, P.t: Phaeodactylum tricornutum, A.f: Asterionella formosa, O.s: Odontella sinensis; F.c: Fragilariopsis cylindrus; Coccolithophores: E.h: Emiliania huxleyi; Other stramenopiles: H.a: Heterosigma akashiwo, N.g: Nannochloropsis gaditana; Rhodophytes: G.l: Gloiocladia laciniata, C.l: Chondria littoralis; Cyanobacteria: S.: Synechococcus sp. PCC 7336, S. sp: Synechocystis sp., A.c: Anabaena cylindrica, A.: Anabaena sp.; Land plants: A.t: Arabidopsis thaliana, S.o: Spinacia oleracea, B.n: Brassica napus, P.m: Prunus mume, S.l: Solanum lycopersicum. Redox-regulated cysteine residues are highlighted by an orange rectangle. Alignments were performed with ClustalW, through MEGA6 software, and processed by GenDoc software.