Figure 2. Screening OB2C small molecule library for death ligands.
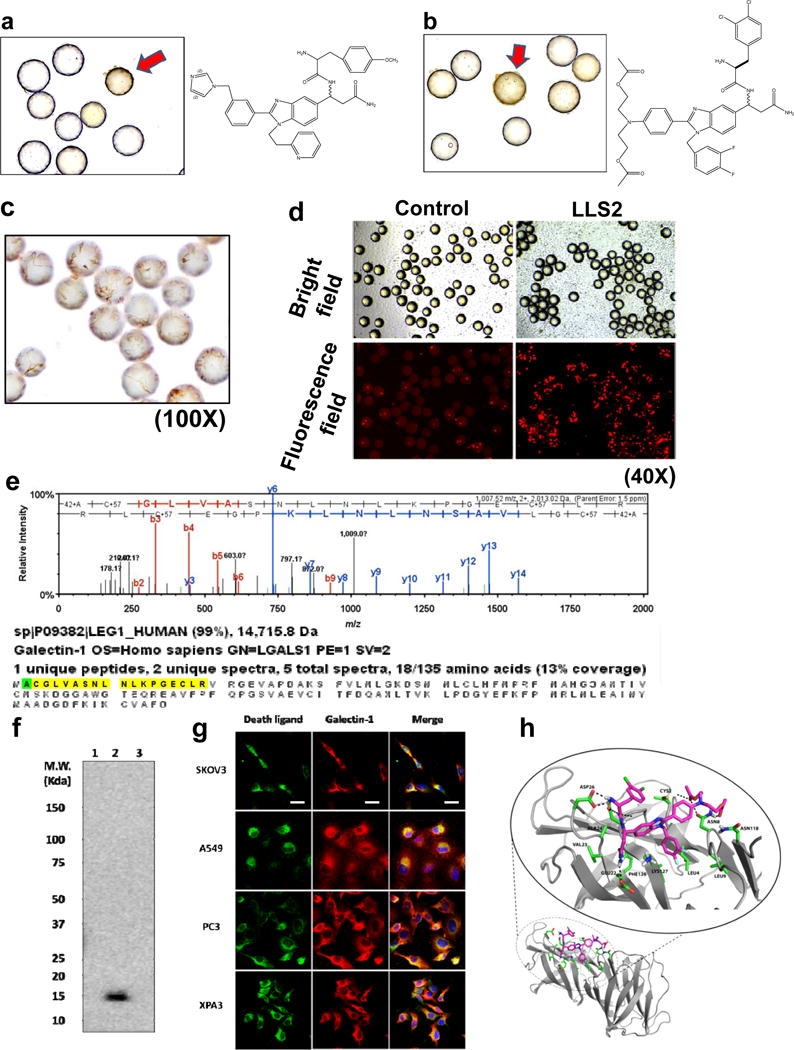
Cleaved caspase 3 positive beads (red arrow) in OB2C beads library. Positive beads were picked up for chemical decoding with automatic Edman microsequencing. (a) LLS1, (b) LLS2. Resynthesize death ligand LLS2 on beads for validation of death effect. (c) Cleaved caspase 3 staining.(100X) (d) Propidium Iodide staining for dead cells. (40X). Galectin-1 is a target protein of LLS2. (e) Eluted protein from pull-down assay was identified as galectin-1 by LC MS/MS. (f) Immunoblot analysis with anti-galectin-1 Ab, to validate the LC MS/MS result. 1: blank bead, 2: LLS2-beasd, 3: a irrelevant small molecule-bead control. (g) Immunostaining result reveals that LLS2 co-localizes with galectin-1. SKOV3, A549, PC3 and XPA3 cells were stained by biotinylated LLS2 (green), galectin-1 antibody (red), and nuclei were stained with DAPI (blue). (h) Computer modeling shows that LLS2 binds the interface of the galectin-1 dimer. Residues that are within 3.5 angstroms from LLS2 were shown. Scale bars: 50 μm.
