Figure 1.
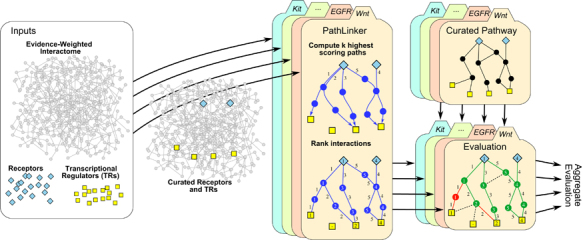
Overview of the PATHLINKER algorithm. Given an interactome, we identify a set of receptors and a set of TRs for a particular curated pathway (e.g., Wnt). We apply PATHLINKER to reconstruct the pathway, ranking proteins and interactions by their first occurrence in the k shortest paths from any receptor to any TR. Using the curated pathway as a ground truth, we evaluate the performance of PATHLINKER. We combine the ranked lists for multiple curated pathways to obtain an aggregate evaluation.
