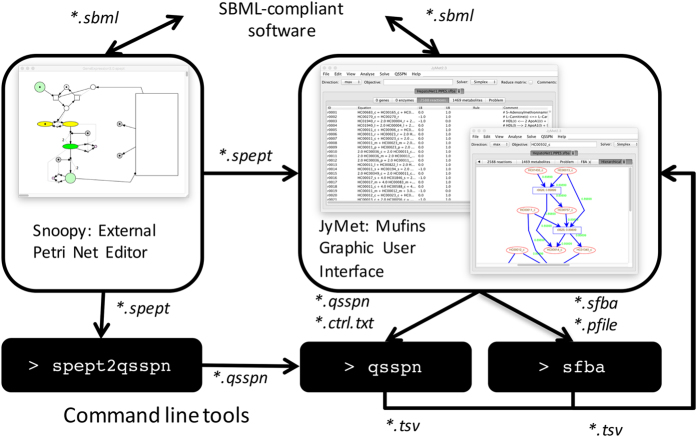
Figure 1.
Overview of MUFINS. All the calculations are performed by two computational engines, which can be also run as stand-alone command line tools. The sfba implements CBM methods and qsspn performs QSSPN simulations. JyMet is a graphic interface to all methods providing spreadsheet representation of models and results as well as metabolic network visualization and plots. JyMet writes input files for computational engines, starts calculations, imports output files and displays results. In the case of QSSPN simulations, Petri Net connectivity can be graphically edited by Snoopy software, a standard Petri Net tool, which we use as external editor. JyMet imports Snoopy files and provides spreadsheet interface allowing editing of QSSPN parameters or independent creation of entire QSSPN model. Conversion of Snoopy files directly to qsspn engine is also possible with command line python script spept2qsspn. Both JyMet and Snoopy import and export SBML file providing connectivity to other SBML-compliant tools. The file formats used for software component communication are indicated by their default extensions and described in Supplementary File Formats. CBM, constraint-based modeling; MUFINS, MUlti-Formalism Interaction Network Simulator; SBML, Systems Biology Markup Language; QSSPN, Quasi-Steady State Petri Net.