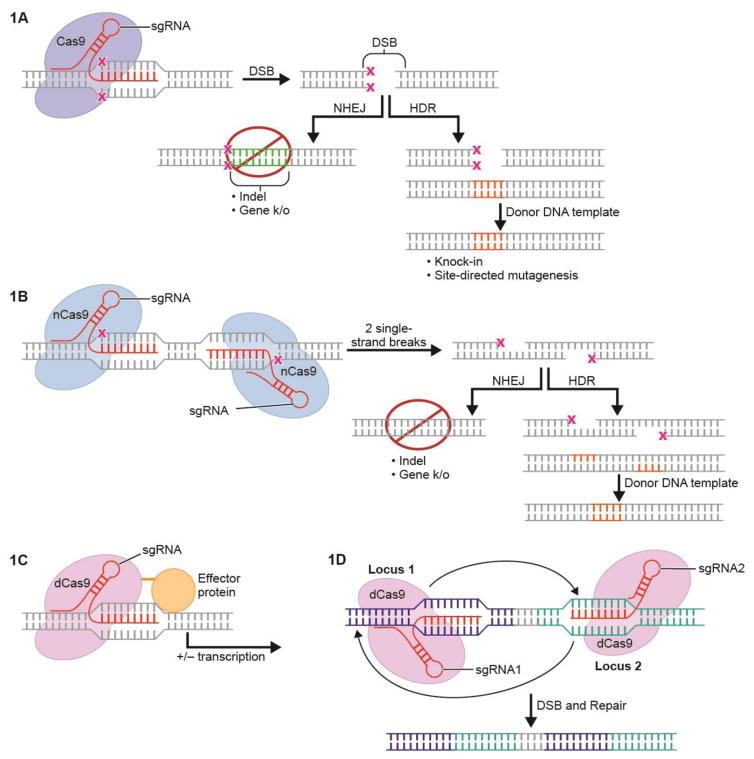
Figure 1. General Applications of CRISPR/Cas9.
A. sgRNA guides Cas9 endonuclease to target locus of interest. A double stranded break (DSB) is made and is repaired in one of two ways. In non-homologous end-joining (NHEJ), DNA is deleted or inserted at the site of the DSB and the gene is knocked out. In homology-directed repair (HDR), a template strand is supplied with homology to the cut region; template DNA is then inserted at the site of the DSB.
B. “nickase” Cas9. Cleavage of one of the two nuclease domains of Cas9 results in nickase Cas9 (nCas9). DSBs are made only at a side in which adjacent single-strand cuts are made by nCas9, each guided by their own sgRNA. A double stranded break is made when single-strand cuts are made at adjacent sites. The DSB can then be repaired with either HDR or NHEJ. This allows for improved specificity of endonuclease-mediated cuts.
C. CRISPR/Cas9 effectors. Fusion of “deactivated” or “dead” (dCas9) to a transcriptional effector allows targeted increase or decrease in the expression of a gene of interest
D. CRISPR/Cas9-mediated genomic rearrangements. DSBs at two sites by two separate sgRNAs can lead to inversion of the region in between the two DSBs.