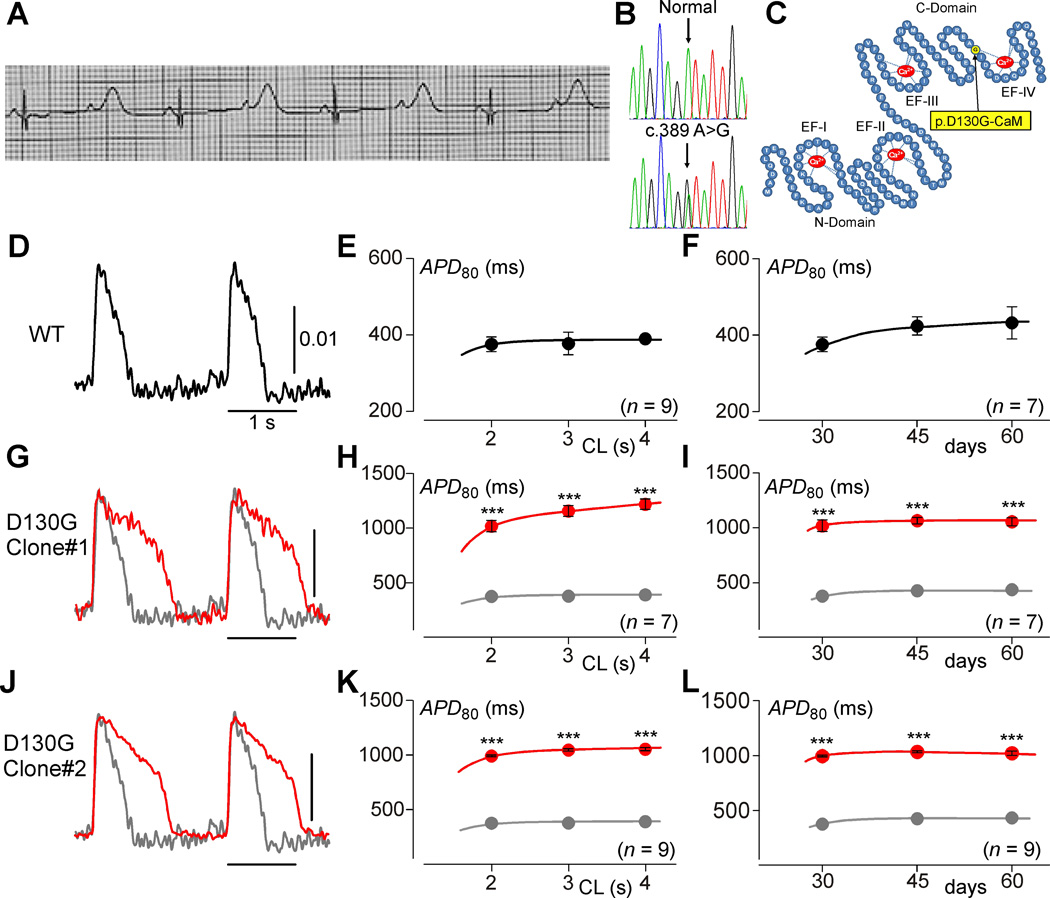
Figure 1. IPSCs recapitulate the calmodulinopathy phenotype.
A, The proband’s electrocardiogram taken at 12 hours after birth, illustrating a QTc>700 ms. B, DNA Sanger sequencing chromatograms for both a normal control (top) and the patient (bottom) revealing a heterozygous c.389 A>G single nucleotide substitution in CALM2 resulting in the p.D130G-CaM amino acid substitution. C, Schematic rendering of the CaM protein (blue) highlighting the N-domain and C-domain, each containing two EF hands (labeled EF-I through EF-IV) with Ca2+ (red) bound. Yellow circle indicates the D130G mutation identified in the LQTS patient. D, Exemplar APs from iPSCWT-CMs (WT) paced at 0.5 Hz, recorded via fluorescence imaging using ASAP1. Scale bar indicates change in fluorescence as measured by ΔF/F0. E, Population data for mean APDs at various pacing cycle lengths (CL) for iPSCWT-CMs (n=9). Error bars indicate ±SEM throughout. Each biological replicate (n) is an average value of 2 technical replicates, here and throughout this figure. F, Population data for iPSCWT-CM APDs at a 2-second cycle length across multiple time points (n=9, 9, and 7 on day 30, 45, and 60, respectively). G, Exemplar APs from iPSCD130G-CaM-CMs (red). WT reproduced in gray. H, Average APD data for iPSCD130G-CaM-CMs (red) (n=7). WT reproduced in gray (*** p<0.001 compared to WT; corrected for unequal variance of normal distributions). I, Average APDs at a 2-second cycle length from D130G iPSCs are stable across time (n=7, 7, and 7 on day 30, 45, and 60, respectively). J–L, Alternate D130G clone demonstrating the same result as G–I (n=9, *** p<0.001 compared to WT and corrected for unequal variance of normal distributions).