Abstract
We have identified a spontaneous mitochondrial mutation, mfs-1 (mitochondrial frameshift suppressor-1), which suppresses a + 1 frameshift mutation localized in the yeast mitochondrial oxi1 gene. The suppressor strain exhibits a single base change (C to U) at position 42 of the mitochondrial serine-tRNA (UCN). To our knowledge, this is the first reported case showing that a mutation in the anticodon stem of a tRNA can cause frameshift suppression. The expression and aminoacylation of the mutant tRNASer(UCN) are not significantly affected. However, the base change at position 42 has two effects: first, residue U27 of the mutant tRNA is not modified to pseudouridine as observed in wild-type tRNASer(UCN). Second, the base change and/or the lack of modification of U27 leads to an alteration in the secondary/tertiary structure of the mutant tRNA. It is possible that there are such structural changes in the anticodon loop that enable the tRNA to read a four base codon, UCCA, thus restoring the wild-type reading frame.
Full text
PDF
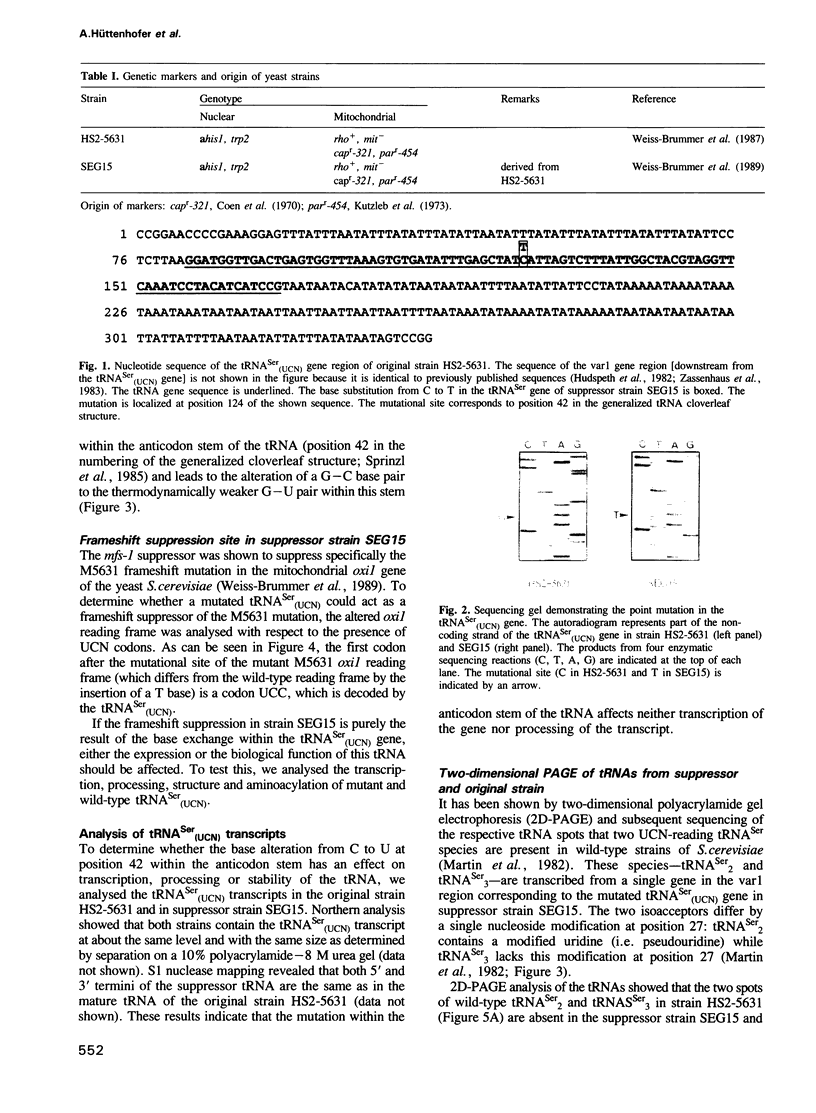
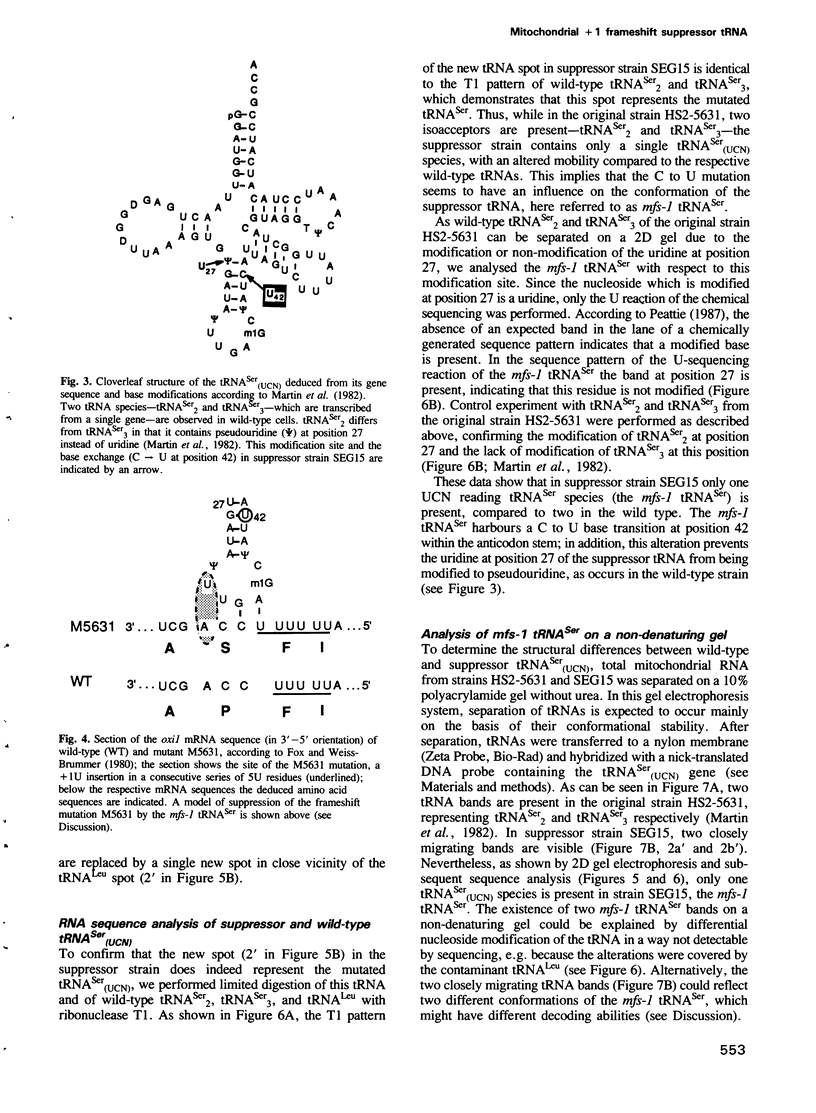




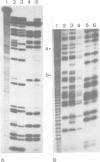
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer D., Yarus M. The context effect does not require a fourth base pair. Science. 1986 Jan 24;231(4736):393–395. doi: 10.1126/science.3510456. [DOI] [PubMed] [Google Scholar]
- Björk G. R., Ericson J. U., Gustafsson C. E., Hagervall T. G., Jönsson Y. H., Wikström P. M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- Bordonné R., Dirheimer G., Martin R. P. Transcription initiation and RNA processing of a yeast mitochondrial tRNA gene cluster. Nucleic Acids Res. 1987 Sep 25;15(18):7381–7394. doi: 10.1093/nar/15.18.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Roth J. R. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell. 1981 Aug;25(2):489–496. doi: 10.1016/0092-8674(81)90067-2. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6105–6109. doi: 10.1073/pnas.81.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D., Deutsch J., Netter P., Petrochilo E., Slonimski P. P. Mitochondrial genetics. I. Methodology and phenomenology. Symp Soc Exp Biol. 1970;24:449–496. [PubMed] [Google Scholar]
- Cummins C. M., Donahue T. F., Culbertson M. R. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3565–3569. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Base substitutions in the tRNA anticodon arm do not degrade the accuracy of reading frame maintenance. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6538–6542. doi: 10.1073/pnas.83.17.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987 Dec 11;238(4833):1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Weiss-Brummer B. Leaky +1 and -1 frameshift mutations at the same site in a yeast mitochondrial gene. Nature. 1980 Nov 6;288(5786):60–63. doi: 10.1038/288060a0. [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):2052–2061. doi: 10.1128/mcb.4.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. The yeast frameshift suppressor gene SUF16-1 encodes an altered glycine tRNA containing the four-base anticodon 3'-CCCG-5'. Gene. 1982 Sep;19(2):163–172. doi: 10.1016/0378-1119(82)90002-6. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Ainley W. M., Shumard D. S., Butow R. A., Grossman L. I. Location and structure of the var1 gene on yeast mitochondrial DNA: nucleotide sequence of the 40.0 allele. Cell. 1982 Sep;30(2):617–626. doi: 10.1016/0092-8674(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Tatti K. M., Grossman L. I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980 Dec 11;610(2):221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- Hughes D., Atkins J. F., Thompson S. Mutants of elongation factor Tu promote ribosomal frameshifting and nonsense readthrough. EMBO J. 1987 Dec 20;6(13):4235–4239. doi: 10.1002/j.1460-2075.1987.tb02772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kutzleb R., Schweyen R. J., Kaudewitz F. Extrachromosomal inheritance of paromomycin resistance in Saccharomyces cerevisiae. Genetic and biochemical characterization of mutants. Mol Gen Genet. 1973 Sep 5;125(1):91–98. doi: 10.1007/BF00292984. [DOI] [PubMed] [Google Scholar]
- Locker J. Analytical and preparative electrophoresis of RNA in agarose-urea. Anal Biochem. 1979 Oct 1;98(2):358–367. doi: 10.1016/0003-2697(79)90154-4. [DOI] [PubMed] [Google Scholar]
- Martin R. P., Schneller J. M., Stahl A. J., Dirheimer G. Study of yeast mitochondrial tRNAs by two-dimensional polyacrylamide gel electrophoresis: characterization of isoaccepting species and search for imported cytoplasmic tRNAs. Nucleic Acids Res. 1977 Oct;4(10):3497–3510. doi: 10.1093/nar/4.10.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Sibler A. P., Dirheimer G. The primary structures of three yeast mitochondrial serine tRNA isoacceptors. Biochimie. 1982 Nov-Dec;64(11-12):1073–1079. doi: 10.1016/s0300-9084(82)80389-1. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Barrell B. G., Seidman J. G. Nucleotide alterations in bacteriophage T4 serine transfer RNA that affect the conversion of precursor RNA into transfer RNA. J Mol Biol. 1975 Dec 25;99(4):717–732. doi: 10.1016/s0022-2836(75)80181-1. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony D. J., Mims B. H., Thompson S., Murgola E. J., Atkins J. F. Glycine tRNA mutants with normal anticodon loop size cause -1 frameshifting. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7979–7983. doi: 10.1073/pnas.86.20.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol. 1973 Apr 25;242(121):230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- Roth J. R. Frameshift suppression. Cell. 1981 Jun;24(3):601–602. doi: 10.1016/0092-8674(81)90086-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong B. L., RajBhandary U. L. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc Natl Acad Sci U S A. 1987 Jan;84(2):334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibler A. P., Dirheimer G., Martin R. P. The primary structure of yeast mitochondrial tyrosine tRNA. FEBS Lett. 1983 Feb 21;152(2):153–156. doi: 10.1016/0014-5793(83)80368-8. [DOI] [PubMed] [Google Scholar]
- Smith D., Yarus M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J Mol Biol. 1989 Apr 5;206(3):489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Moll J., Meissner F., Hartmann T. Compilation of tRNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r1–49. doi: 10.1093/nar/13.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S. D., Murgola E. J., Pagel F. T. Missense and nonsense suppressors can correct frameshift mutations. Biochimie. 1989 Jun;71(6):729–739. doi: 10.1016/0300-9084(89)90089-8. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Hüttenhofer A. The paromomycin resistance mutation (parr-454) in the 15 S rRNA gene of the yeast Saccharomyces cerevisiae is involved in ribosomal frameshifting. Mol Gen Genet. 1989 Jun;217(2-3):362–369. doi: 10.1007/BF02464905. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Sakai H., Hüttenhofer A. A mitochondrial frameshift suppressor maps in the tRNASer-var1 region of the mitochondrial genome of the yeast S. cerevisiae. Curr Genet. 1989 Apr;15(4):239–246. doi: 10.1007/BF00447038. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Sakai H., Kaudewitz F. A mitochondrial frameshift-suppressor (+1) [corrected] of the yeast S. cerevisiae maps in the mitochondrial 15S rRNA locus. Curr Genet. 1987;11(4):295–301. doi: 10.1007/BF00355403. [DOI] [PubMed] [Google Scholar]
- Weiss R., Gallant J. Mechanism of ribosome frameshifting during translation of the genetic code. 1983 Mar 31-Apr 6Nature. 302(5907):389–393. doi: 10.1038/302389a0. [DOI] [PubMed] [Google Scholar]
- Winey M., Mendenhall M. D., Cummins C. M., Culbertson M. R., Knapp G. Splicing of a yeast proline tRNA containing a novel suppressor mutation in the anticodon stem. J Mol Biol. 1986 Nov 5;192(1):49–63. doi: 10.1016/0022-2836(86)90463-8. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Farrelly F., Hudspeth M. E., Grossman L. I., Butow R. A. Transcriptional analysis of the Saccharomyces cerevisiae mitochondrial var1 gene: anomalous hybridization of RNA from AT-rich regions. Mol Cell Biol. 1983 Sep;3(9):1615–1624. doi: 10.1128/mcb.3.9.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]