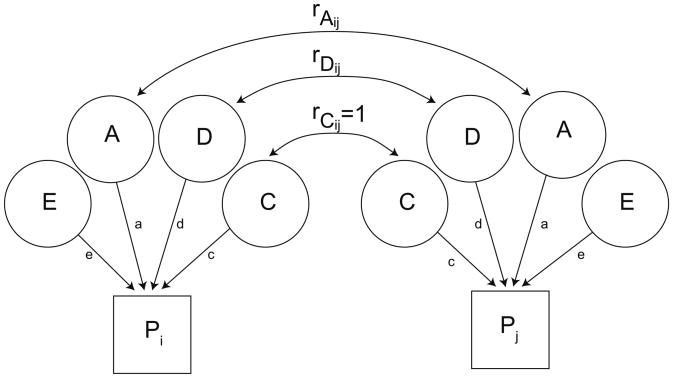
Fig. 2.
A schematic of the biometric latent variable model. Two family members (individuals i and j) are included to illustrate the model. Their phenotypes are represented by the boxes labeled Pi and Pj. Each phenotype is due to the influence of the four latent variables (ADCE). The variance of each family member's phenotype and the covariance between each pair of family members are determined by the path estimates from each latent variable to the phenotype (a, d, c, and e) and the covariance between family members. The variances of each latent variable are fixed at 1. Their covariances are a function of the family relationships. The genetic covariance between pairs of individuals is depicted as rAij and rDij for additive and dominance variance, respectively. It equals 1 for MZ twins for both types of inheritance, whereas the covariance due to A is ½ for DZ twins and parent-offspring pairs. The covariance due to D is ¼ for DZ twins. C is assumed the same for all pairs and is equal to 1. E is by definition unique to each individual (uncorrelated for pairs of family members).