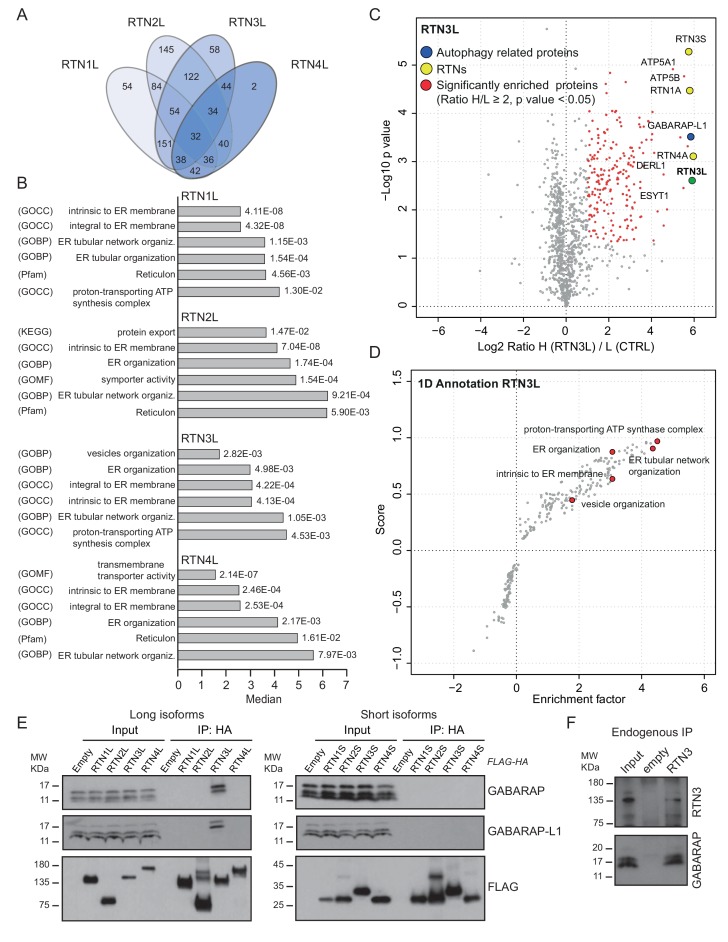
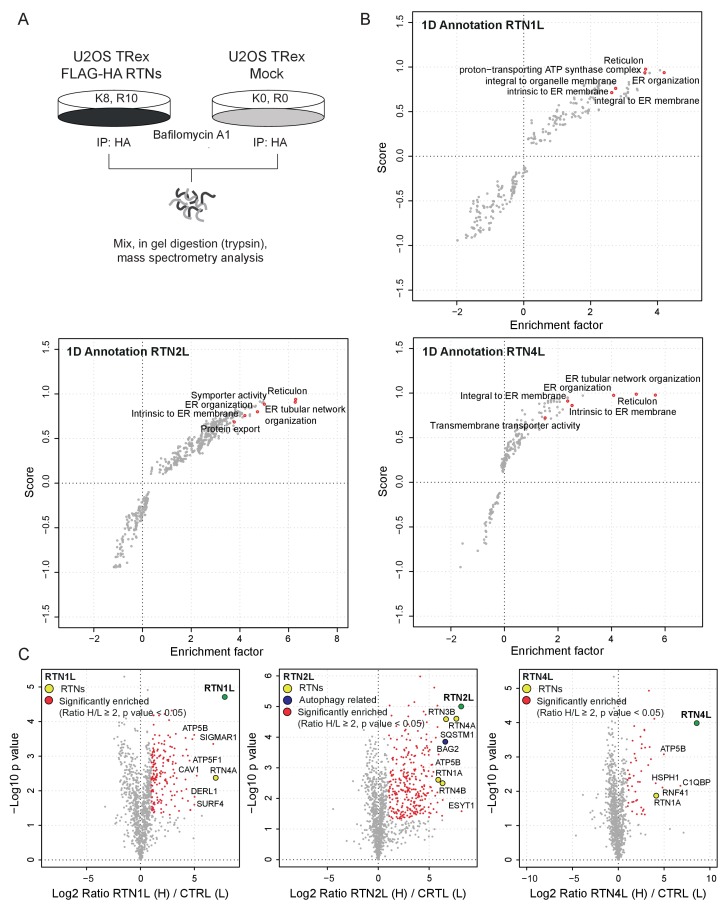
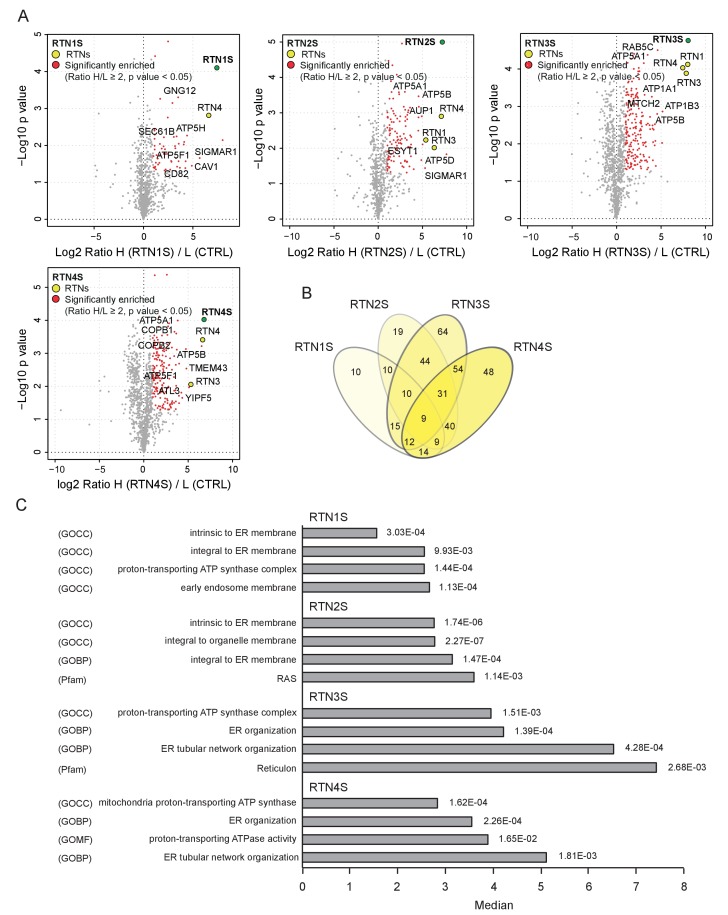
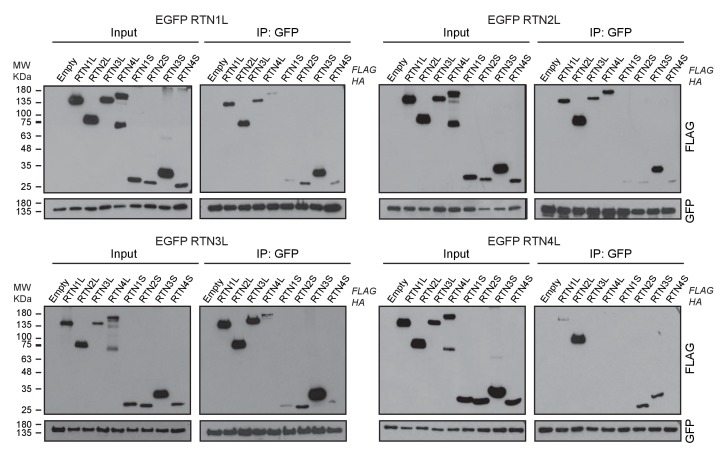
Figure 5. RTN3 interacts with the autophagy modifiers.
(A) Venn diagrams of the interactors of the four RTNs. Numbers represent the identified peptides significantly enriched in three IP and mass spectrometry replicates for each RTN. (B) Annotation enrichment analysis of the interactors of long RTN1-4 isoforms. Bars represent the significantly enriched gene ontology biological process (GOBP), the gene ontology cellular components (GOCC), the gene ontology molecular function (GOMF), the over-expressed pathways (KEGG) and the domain enrichment (Pfam). The numeric value on the right side of the bar shows the Benjamini-Hochberg FDR value. (C) Scatter-plot for 1D annotation enrichment analysis of RTN3L interactor partners significantly enriched in three different IPs. (D) Volcano-plot for RTN3L SILAC-based interactome. Peptides with and Log2 Ratio H/L ≥1 and –Log10 p value > 1.3 are labeled in red. Three biological replicates were analyzed. (E) Co-IP of endogenous GABARAP and GABARAP-L1 with over-expressed long and short isoforms of RTN1-4. Over-expression was induced for 24 hr in U2OS TRex stable cell lines using 1 µg/ml of doxycycline. Bafilomycin A1 was added at the final concentration of 200 ng/ml for 2 hr. (F) Endogenous Co-IP of RTN3 with GABARAP in A549 cells. The ‘empty’ lane represents unconjugated beads. Bafilomycin A1, 200 ng/ml, was added for 2 hr.
DOI: http://dx.doi.org/10.7554/eLife.25555.019