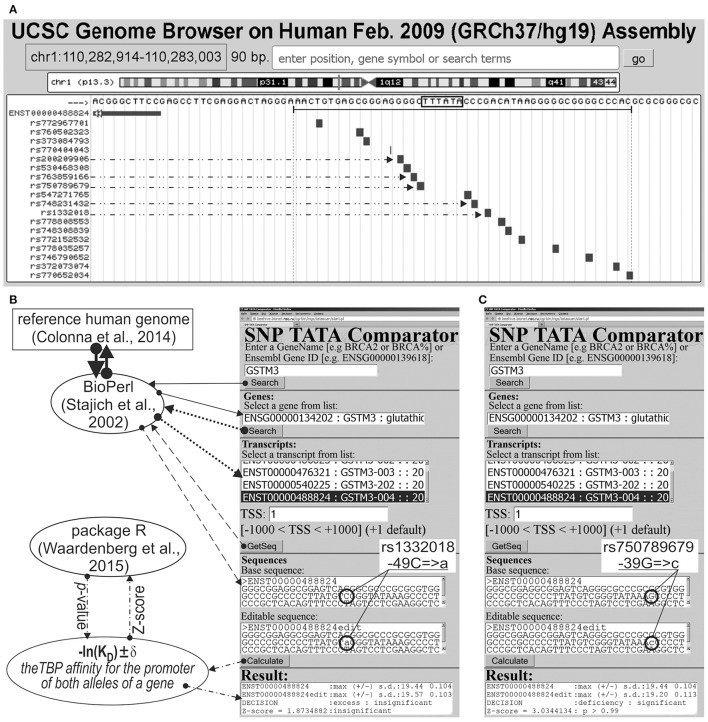
Figure 1.
Known and candidate SNP markers of Alzheimer's disease (AD) near TBP-binding sites of the human GSTM3 gene promoter. (A) Unannotated SNPs (analyzed in this study) in the region [−70; −20] (where all proven TBP-binding sites (boxed) are located; double-headed arrow, ↔) of the human GSTM3 gene promoter taken from dbSNP (Sherry et al., 2001) using the UCSC Genome Browser (Haeussler et al., 2015). Dash-and-double-dot arrows: known and candidate SNP markers of sporadic AD are predicted by a significant change in the affinity of TBP for the human GSTM3 gene promoter. (B,C) The results from our Web service (Ponomarenko et al., 2015) for the two SNP markers of sporadic AD: known rs1332018 (Hong et al., 2009; Tan et al., 2013) and candidate marker rs750789679 near the known TBP-binding site (boxed) of the human GSTM3 gene promoter. Solid, dotted, and dashed arrows indicate queries for the gene list, list of transcripts of a certain gene, and DNA sequence of the promoter corresponding to the specified transcript by means of the BioPerl library (Stajich et al., 2002) of the reference human genome (Colonna et al., 2014), respectively. Dash-and-dot arrows: estimates of significance of the alteration of gene product abundance in patients with the minor allele (mut) relative to the norm (ancestral allele, wt) expressed as a Z-score using package R (Waardenberg et al., 2015). Circles indicate the ancestral (wt) and minor (mut) alleles of the SNP marker labeled by its dbSNP ID (Sherry et al., 2001).