Figure 1.
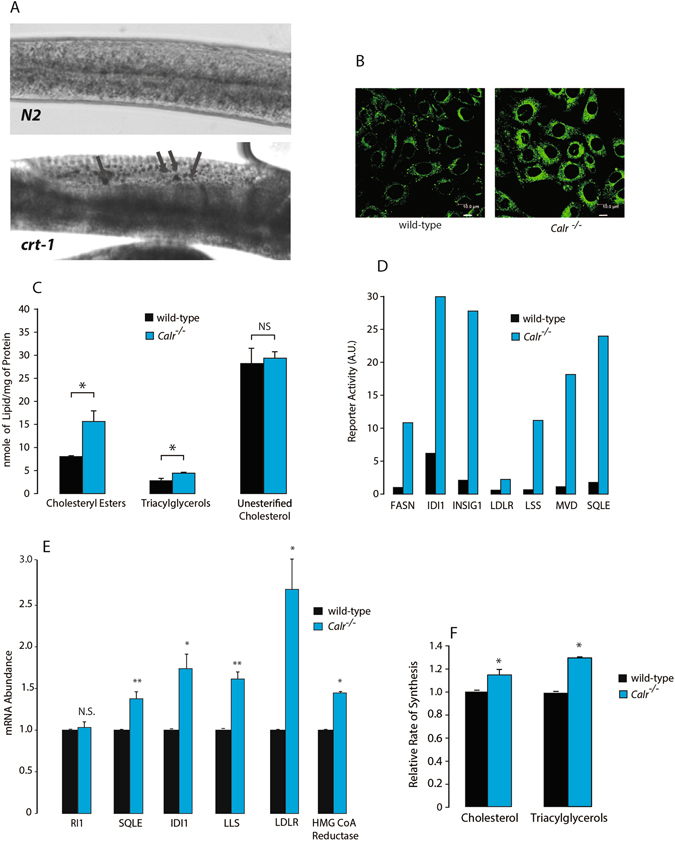
Lipids in the absence of calreticulin. (A) Sudan black staining of neutral lipids in wild-type (N2) and crt-1 calreticulin deficient (jh101) C. elegans. Lipid droplets are indicated by the arrows. (B) BODIPY 505/515 staining of neutral lipids in wild-type and calreticulin-deficient (Calr −/−) cells. (C) Cholesteryl esters, triacyglycerols and unesterified cholesterol levels in wild-type and Calr −/− cells. *Indicates statistically significant differences: cholesteryl esters, p-value = 0.0374; triacylglycerols, p-value = 0.0451. NS, not significant. Representative of 3 biological replicates. Student’s t-test was used. (D) SwitchGear Cholesterol Biosynthesis Pathway assay for SREBP target genes in wild-type and Calr −/− cells; fatty acid synthase (FASN), isopentenyl-diphosphate delta isomerase (IDI1), insulin induced gene-1 (INSIG1), low density lipoprotein receptor (LDLR), lanosterol synthase (LSS), mevalonate pyrophosphate decarboxylase (MVD) and squalene epoxidase (SQLE) Representative of 2 biological replicates. (E) Q-PCR analysis of replication initiator 1 (RI1), squalene (SQLE), isopentenyl-diphosphate delta isomerase (IDI1), lanosterol synthase (LSS), LDL receptor (LDLR) and HMG-CoA reductase mRNA level in wild-type and Calr −/− cells. Results were normalized to 18S rRNA (internal control). **Indicates statistically significant differences: SQLE in wild-type vs. Calr −/− cells, p-value = 0.0041; LSS in wild-type vs. Calr −/− cells, p-value = 0.001. *Indicates statistically significant differences: IDI1 in wild-type vs. Calr −/− cells, p-value = 0.0133; LDLR in wild-type vs. Calr −/− cells, p-value = 0.0112; HMG-CoA Reductase in wild-type vs. Calr −/− cells, p-value = 0.0001. Representative of 4 biological replicates. NS, not significant (Student’s t-test). (F) Analysis of rates of synthesis of cholesterol and triacyglycerols in wild-type and Calr −/− cells. *Indicates statistically significant differences: cholesterol, p-value = 0.0012; triacylglycerols, p-value = 0.0006 (Student’s t-test). See “Experimental Procedures” for additional details.
