Table 2.
Literature summary of molecular studies comparing sinus bacteriology of patients with and without CRS
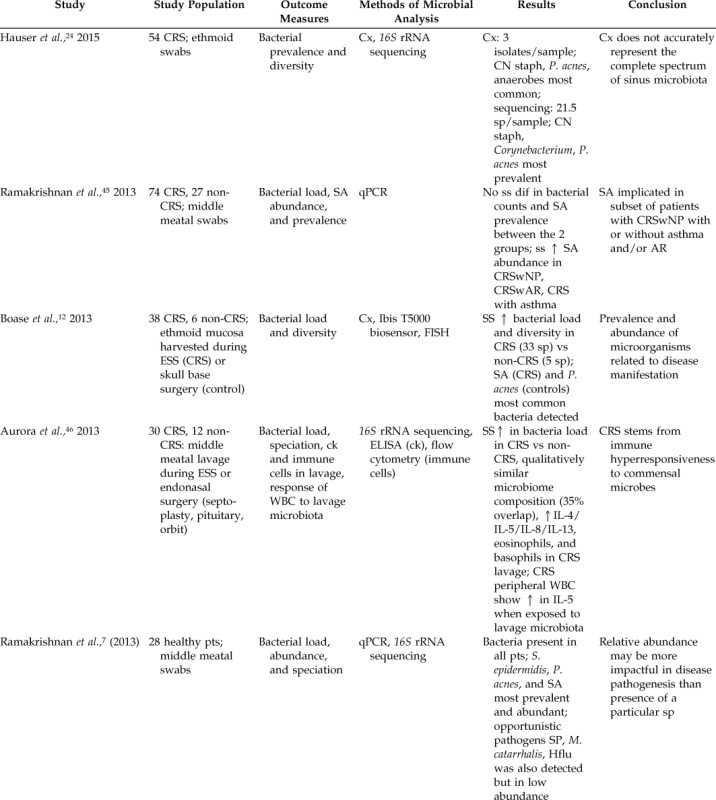
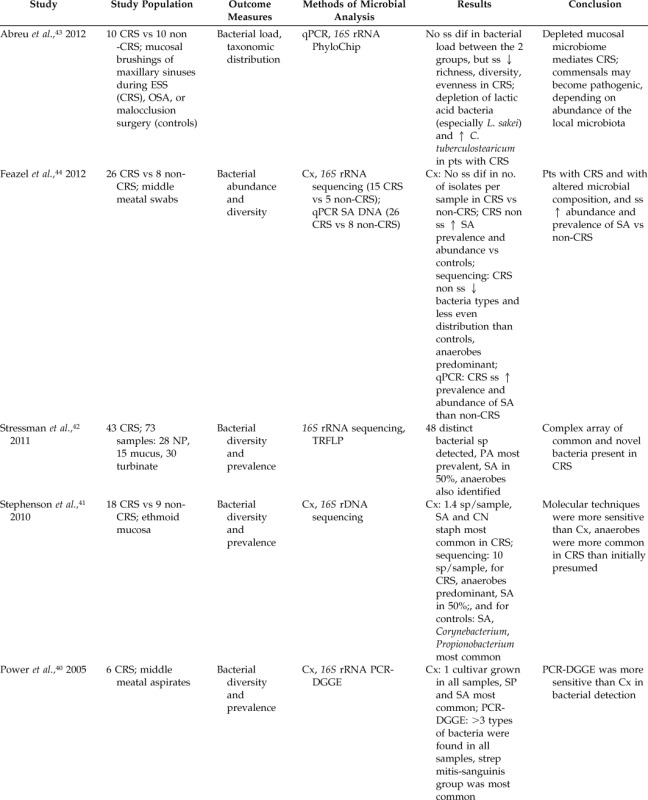
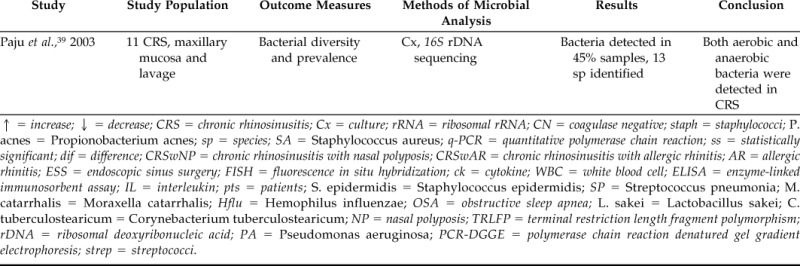
↑ = increase; ↓ = decrease; CRS = chronic rhinosinusitis; Cx = culture; rRNA = ribosomal rRNA; CN = coagulase negative; staph = staphylococci; P. acnes = Propionobacterium acnes; sp = species; SA = Staphylococcus aureus; q-PCR = quantitative polymerase chain reaction; ss = statistically significant; dif = difference; CRSwNP = chronic rhinosinusitis with nasal polyposis; CRSwAR = chronic rhinosinusitis with allergic rhinitis; AR = allergic rhinitis; ESS = endoscopic sinus surgery; FISH = fluorescence in situ hybridization; ck = cytokine; WBC = white blood cell; ELISA = enzyme-linked immunosorbent assay; IL = interleukin; pts = patients; S. epidermidis = Staphylococcus epidermidis; SP = Streptococcus pneumonia; M. catarrhalis = Moraxella catarrhalis; Hflu = Hemophilus influenzae; OSA = obstructive sleep apnea; L. sakei = Lactobacillus sakei; C. tuberculostearicum = Corynebacterium tuberculostearicum; NP = nasal polyposis; TRLFP = terminal restriction length fragment polymorphism; rDNA = ribosomal deoxyribonucleic acid; PA = Pseudomonas aeruginosa; PCR-DGGE = polymerase chain reaction denatured gel gradient electrophoresis; strep = streptococci.
