Fig. 2.
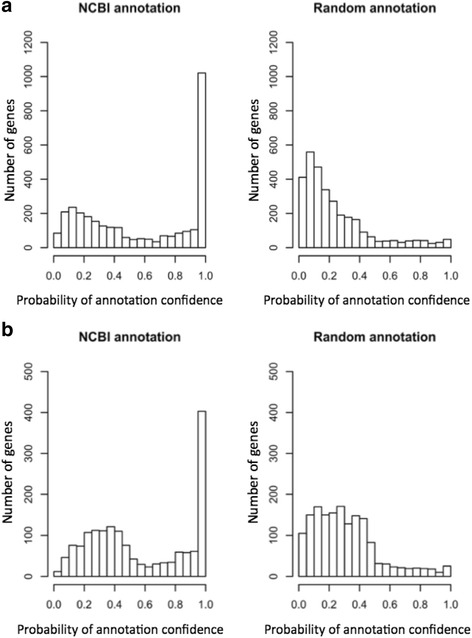
Distributions of PAC values of NCBI and random annotations for (a) E. coli and (b) C. thermocellum. a Using conditional probabilities derived from a given genome and observed gene function similarities, we calculated PAC values for NCBI annotation (assumed to be correct) and random annotation (assumed to be incorrect) for the E. coli strain K-12 substrain MG1655. The probability bin [0.95, 1] has 1021 genes for NCBI annotation and 49 genes for random annotation of 3117 genes applicable to PAC calculation. b We applied the same methodology to C. thermocellum. The probability bin [0.95, 1] contains 403 genes for NCBI annotation and 25 genes for random annotation among 1617 genes applicable to PAC calculation
