Fig. 2.
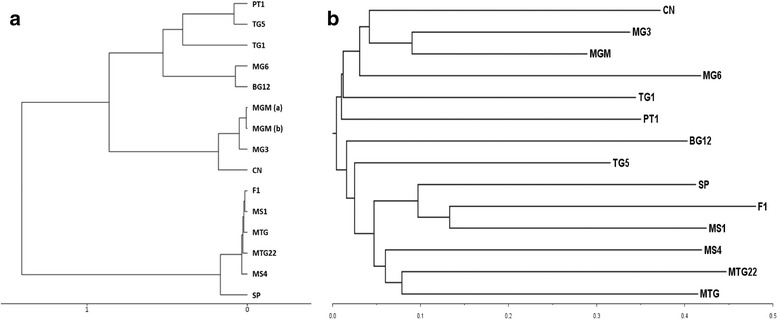
Reference-independent-based clustering of Antarctic viromes. a Hierarchical clustering based on dinucleotide frequencies in the contigs calculated by the MetaVir server pipeline. The x-axis denotes eigenvalues distances. Virome read datasets were assembled to contigs prior to analysis. Dataset MGM was split in two parts (a, b) due to size limitations of the pipeline. b crAss clustering of the 14 viromes in this study. The distances between viromes were calculated by crAss [34] using the SHOT distance formula [71], while the cladogram was created by crAss with BioNJ [72] and visualized with FigTree [73]. The branch lengths and scale axis represent the distance measures
