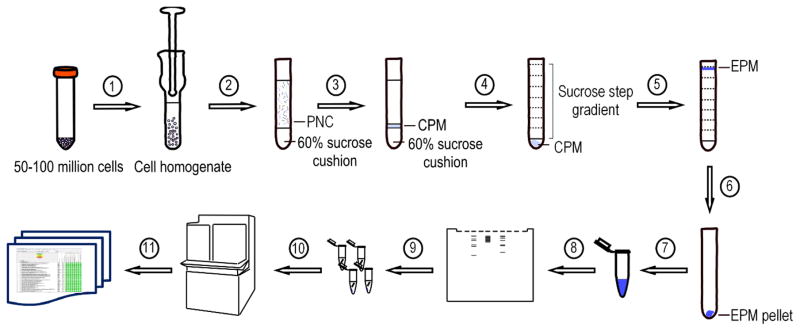
Figure 1.
Experimental workflow for the proposed label-free, non-affinity-enriched surfaceome protocol. (1) Resuspend cells in buffer and homogenize with a Dounce homogenizer. Spin homogenate at 1,000g for 10 minutes at 4°C. (2) Layer the PNC over a 60% sucrose cushion and centrifuge at 100,000g for an hour at 4°C. (3) Recover the CPM fraction from the top of the sucrose cushion and transfer to a centrifuge tube. (4) Layer sucrose step gradient on top of the CPM. Centrifuge at 100,000g overnight at 4°C. (5) Recover the EPM fraction at the 37% sucrose fraction border phase and transfer to a centrifuge tube. (6) Fill the tube with HEPES buffer and centrifuge at 150,000g for an hour 4°C. (7) Dissolve the EPM pellet in 300 μL ammonium bicarbonate buffer. Acetone precipitate all or part of the protein sample. (8) Denature protein sample and run on an SDS-PAGE. (9) Cut the gel in thin slices and perform in-gel trypsin digestion. (10) Extract the tryptic peptides and analyze by LC-MS/MS. (11) Bioinformatic data processing and analysis.
PNC, post-nuclear cytosol; CPM, crude plasma membrane; EPM, enriched plasma membrane