Fig. 4.
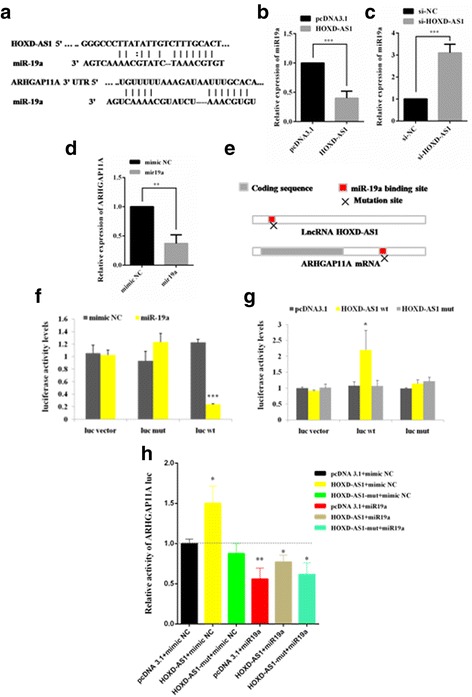
HOXD-AS1 interacts with miR-19a and controls ARHGAP11A expression levels. a The prediction for miR-19a binding sites on the HOXD-AS1 transcript and the potential target gene ARHGAP11A. b and c Relative expression levels of miR19a (compared to U6) after pcDNA3.1-HOXD-AS1 or siRNA treatment measured by QRT-PCR. d Relative expression levels of ARHGAP11A (compared to GAPDH) after miR19a or mimic treatment measured by QRT-PCR. e Schematic outlining the predicted binding sites and mutation site of miR19a on HOXD-AS1 and the ARHGAP11A transcript. f Luciferase activity in HCCLM3 cells cotransfected with miR-19a and empty luciferase reporter or vectors containing the wildtype (luc wt) 3'-UTR region of the ARHGAP11A transcript or mutant (luc wt) transcript. g Luciferase activity in HCCLM3 cells cotransfected with wildtype (wt) or mutant (mut) HOXD-AS1 overexpression plasmids and empty luciferase reporter or vectors containing the wildtype (luc wt) 3'-UTR region of the ARHGAP11A transcript or mutant (luc mut) transcript. h Luciferase activity in HCCLM3 cells transfected with ARHGAP11A 3'-UTR luciferase reporter vectors, luc wt, luc mut or empty vectors. f-h The data are presented as the relative ratio of firefly luciferase activity to renilla luciferase activity. The data are shown as the mean ± SD. Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001
