Figure 4.
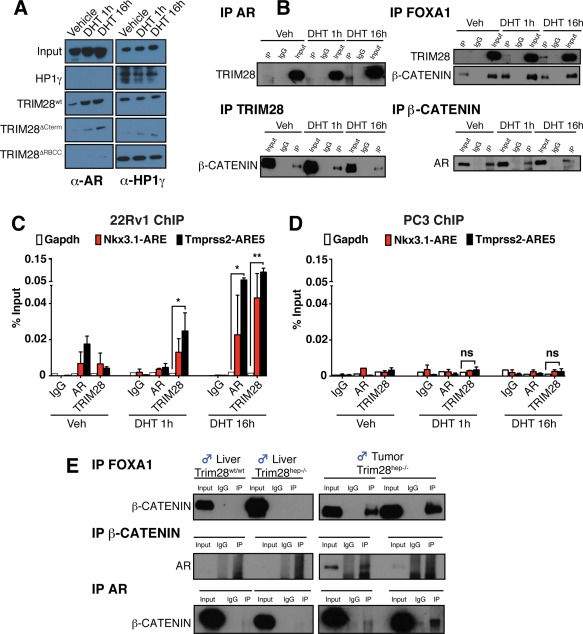
Biochemical characterization of TRIM28‐associating partners. (A) Purified recombinant wt or GST‐TRIM28 deletion mutants were incubated with 22Rv1 cell lysates, and the pull‐down complexes were analyzed by western blotting with the indicated antibodies. As a positive control, pull‐down complexes were incubated with HP1γ antibody. Recombinant GST‐HP1γ was used as negative control for AR interaction. n = 4. (B) IPs were performed using nuclear extracts (NEs) prepared from methanol‐stimulated (Veh) or DHT‐stimulated 22Rv1 cells using antibodies against TRIM28, β‐CATENIN, FOXA1, and AR. IP complexes were analyzed by western blotting using indicated antibodies. (C,D) 22Rv1 (C) or PC3 (D) cells were stimulated for 1 or 16 hours with DHT or vehicle alone (Veh). Chromatin was immunoprecipitated with antibodies recognizing TRIM28, AR, and IgG as a negative control and quantitated by qPCR using primer sets specific for indicated genomic regions. Gapdh promoter region was used as negative control. Values are represented as mean ± SEM as a percentage of the input chromatin; n = 3, *P < 0.05; **P < 0.01; ns (not significant), by two‐way ANOVA followed by Bonferroni's posttest. (E) In vivo IPs were performed on protein extract from livers and tumors of Trim28wt/wt and Trim28hep–/– mice incubated with antibodies against FOXA1 (top panels), β‐CATENIN (middle panels), and AR (bottom panels), and the resulting eluates were analyzed by western blotting with the indicated antibodies. Abbreviations: Gapdh, glyceraldehyde‐3‐phosphate dehydrogenase; HP1γ, heterochromatin protein 1‐gamma; IgG, immunoglobulin G; IPs, immunoprecipitations; Veh, vehicle.
