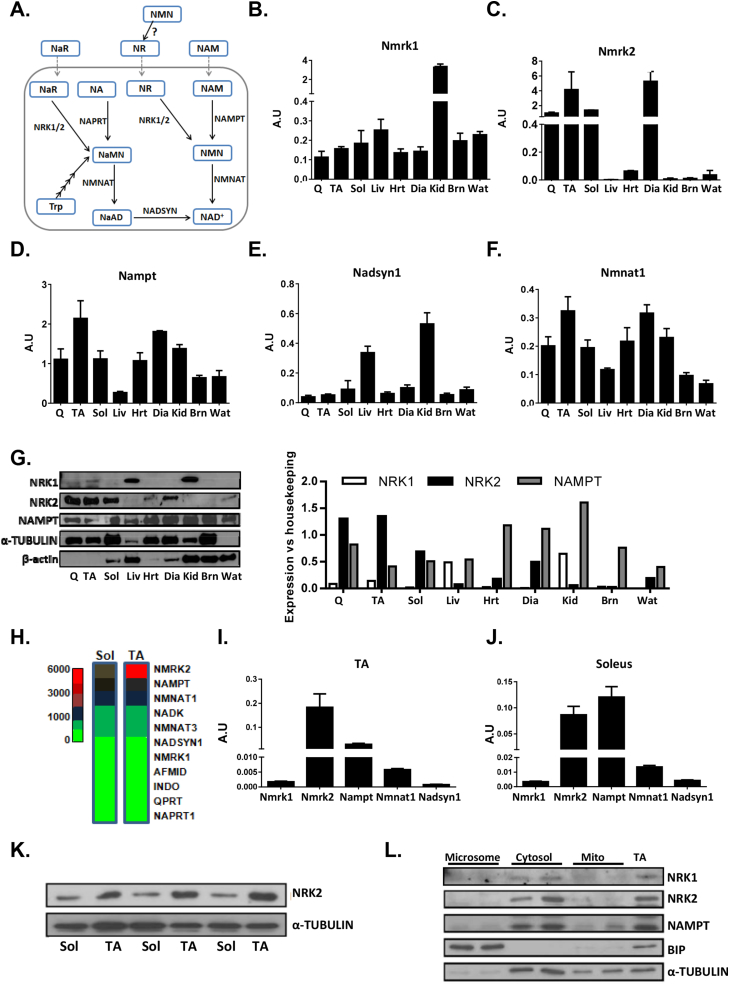
Figure 1.
NAD+biosynthesis pathways in skeletal muscle. (A) An illustration of the known NAD+ biosynthesis pathways identified in mammalian cells with exogenous precursor supplementation strategies illustrated with dashed lines. NAD+ can be synthesized de novo from tryptophan (Trp) and following multiple enzymatic reactions is converted to nicotinic acid mononucleotide (NaMN) and then nicotinic acid dinucleotide (NaAD) by nicotinamide mononucleotide adenylyltransferase (NMNAT) activity, before the final conversion to NAD+ by NAD synthase (NADSYN). NAD+ can be salvaged from precursors nicotinic acid riboside (NaR) and nicotinic acid (NA) by nicotinamide riboside kinases (NRK1/2) and nicotinic acid phosphoribosyltransferase (NAPRT) respectively to NaMN and follow the same final steps. Finally, nicotinamide riboside (NR) and nicotinamide (NAM) are salvaged by NRKs and nicotinamide phosphoribosyltransferase (NAMPT) respectively to nicotinamide mononucleotide (NMN) and finally to NAD+ by NMNAT activity. Real-time PCR mRNA expression of Nmrk1(B), Nmrk2(C), Nampt(D), Nadsyn1 (E), and Nmnat1 (F) across metabolic tissues. (G) Protein expression of NRK1, NRK2, and NAMPT across metabolic tissues with corresponding densitometry compared to housekeeping protein (α-Tubulin for all tissues except liver and Wat which are compared to β-actin expression). (H) Microarray of NAD+ biosynthesis genes in tibialis anterior (TA) and soleus (Sol) skeletal muscle tissue (n = 3). Real-time PCR mRNA expression of rate limiting NAD biosynthesis genes in TA (I) and soleus (J) muscle (n = 4). (K) NRK2 protein expression in TA and soleus muscle tissue. (L) NRK1, NRK2, and NAMPT protein expression in cytosolic, mitochondrial and microsomal fractions of skeletal muscle. (Q = quadriceps, TA = tibialis anterior, Sol = soleus, Liv = liver, Hrt = heart, Dia = diaphragm, Kid = kidney, Brn = brain and Wat = white adipose tissue).