Abstract
Purpose
To reveal the underlying genetic defect in two four-generation Chinese families with aniridia and explore the pathologic mechanism.
Methods
Full ophthalmic examinations were performed in two families with aniridia. The PAX6 gene was directly sequenced in patients of two families, and the detected variants were screened in unaffected family members and two hundred unrelated healthy controls. Real-time quantitative PCR was used to explore pathologic mechanisms of the two variants.
Results
Aniridia, cataract, and oscillatory nystagmus were observed in patients of the two families. In addition, we observed corneal opacity and microphthalmus in family 1, and strabismus, left ectopia lentis, microphthalmus, and microcornea in family 2. Sanger sequencing detected a novel 1-bp duplication (c.50dupA) in family 1 and a novel 2-bp splice site deletion (c.765+1_765+2delGT) in family 2. Sequencing of cDNA indicated skipping of exon 9 caused by the splice site deletion, being predicted to cause a premature stop codon, as well as the duplication. The PAX6 mRNA significantly lower in patients with aniridia than in unaffected family members in both families, suggesting that the duplication and splice site deletion caused nonsense-mediated mRNA decay.
Conclusions
Our study identified two novel PAX6 variants in two families with aniridia and revealed the pathogenicity of the variants; this would expand the variant spectrum of PAX6 and help us better understand the molecular basis of aniridia, thus facilitating genetic counseling.
Introduction
Aniridia (OMIM 106210) is a rare but severe congenital ocular disorder with an incidence of approximately one in 64 000 to 96 000 live births worldwide.1, 2 In addition to incomplete formation of the iris, other clinical manifestations, including cataract, nystagmus, corneal opacity, glaucoma, lens dislocation, and foveal hypoplasia with obvious loss of vision, have been found in aniridia patients.3, 4 Approximately two-thirds of cases are familial with an autosomal dominant inheritance pattern, complete penetrance and variable expressivity, and the remaining third are sporadic.5 A small number of the patients also suffer from WAGR syndrome (Wilms tumor, Aniridia, Genitourinary anomalies and mental Retardation), which is caused by deletion of chromosome 11p13 involving WT1 and PAX6 genes.6
The human paired box gene 6 (PAX6) is a member of the paired box gene families and encodes the PAX6 transcription factor, spanning more than 22 kb at chromosome 11p13. PAX6 gene contains 14 exons and generates 3 transcript isoforms by alternative splicing.7 The PAX6 protein consists of 3 domains: a paired domain (PD) in the NH2-terminal region, a homeodomain (HD), and a proline-serine-threonine domain (PST) in the COOH-terminal terminus.8 The homozygous loss of PAX6 is usually lethal to the embryo in early gestation. To date, 433 PAX6 heterozygous variants have been identified and recorded in the Human PAX6 Allelic Variant Database (Leiden Open Variation Database, LOVD), and more than three quarters of which are frameshift variants.9 Tzoulaki et al speculated that the aniridia phenotype is predominantly associated with variants that introduce a premature termination codon (PTC), whereas non-aniridia phenotypes are typically missense variants.10, 11 In the current study, we screened all PAX6 exons and their adjacent splicing junctions in two Chinese families with severe clinical manifestations including aniridia, cataract, and nystagmus, and identified a novel 1-bp duplication and a novel 2-bp splice site deletion, both leading to PTCs and triggered nonsense-mediated mRNA decay (NMD).
Materials and methods
Subject recruitment and clinical examination
Two families with congenital aniridia were recruited and identified at the Hunan Jiahui Genetics Hospital. Four members of family 1 (two affected and two unaffected, one male and three females) and four members of family 2 (two affected and two unaffected, two males and two females) took part in this study. Visual acuity, refraction error, intraocular pressure, ocular fundus, and slit lamp examinations were performed and documented for all participants. Two hundred randomly selected normal individuals were included as controls. None of the controls had any history of ophthalmic disease. The study was approved by the Ethics Committee of Hunan Jiahui Genetics Hospital. Informed consent was signed by each of the subjects.
Genomic DNA extraction and variant analysis
Genomic DNA of all subjects was extracted from peripheral blood cells by phenol/chloroform method. All coding regions and intron/exon boundaries of PAX6 gene were amplified by PCR. PCR primers were designed using PRIMER5 software (PREMIER Biosoft International, Palo Alto, CA, USA). PCR products were verified by polyacrylamide gel electrophoresis (PAGE) and directly sequenced with an ABI PRISM BigDye kit on an ABI 3130 DNA sequencer (Applied Biosystems, Carlsbad, CA, USA). Sequences were analyzed using the DNASTAR package (DNASTAR, Madison, WI, USA). Two hundred unrelated healthy controls were also screened to identify possible polymorphisms.
RNA extraction and reverse transcription PCR
Total RNA was isolated from cultured lymphocyte cell lines from the proband and unaffected family members using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as indicated in the manufacturer’s protocol. One microgram of total RNA from each sample was reverse transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Carlsbad, CA, USA). Two primers were designed based on the cDNA sequence in order to explore the activation of new splice site in family 2. The primer sequences were 5′-CATCCTTTACCCAAGAGCAAATT-3′ (forward) and 5′-TGGTGCTGAAACTACTGCTGATAG-3′ (reverse). After PCR, the products were verified by agarose gel electrophoresis and sequenced directly.
Real-time quantitative PCR analysis
To uncover expression patterns of the variants, we performed real-time quantitative PCR (RT-qPCR) on the cDNA of the proband and unaffected family members in both families with the Maxima SYBR Green qPCR Master Mix (Thermo Fisher, Carlsbad, CA, USA). Three replicates of RT-qPCR experiments were performed using a Bio-Rad CFX96 Touch Deep Well Real-Time PCR Detection System (Bio-Rad, Berkeley, CA, USA). The primer sequences used were 5′-GATTACTGTCCGAGGGGGTCT-3′ (forward) and 5′-CCTTAGTTTATCATACATGCCGTCT-3′ (reverse). The β-actin gene was employed as a reference with the primers 5′-CACGATGGAGGGGCCGGACTCATC-3′ (forward) and 5′-TAAAGACCTCTATGCCAACACAGT-3′ (reverse).
Results
Clinical findings
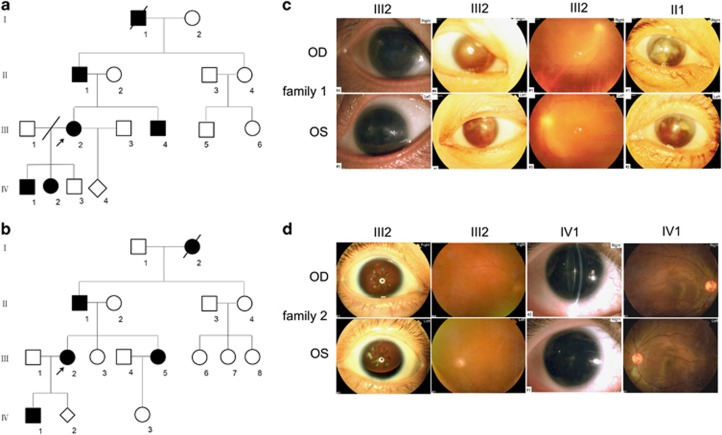
As illustrated in Table 1 and Figure 1, four affected patients in two four-generation families receiving ophthalmic examination showed similar clinical symptoms (Figures 1a and b). In family 1, the proband showed aniridia, cataract, oscillatory nystagmus, corneal opacity, and microphthalmus. Her visual acuities were both 0.05. Her father presented even more severe symptoms, including being totally blind, ocular fundus examination failure resulting from the lens opacity and corneal pannus (Figure 1c). In family 2, the proband showed aniridia, cataract, oscillatory nystagmus, strabismus, and left ectopia lentis. The son of the proband presented microphthalmus, microcornea, weakened foveal reflex, and residual iris in his left eye, with complete aniridia in the other (Figure 1d). He exhibited vitreous opacity and scattered punctate opacities in the lens cortex and capsule. No lens opacity was identified by ophthalmic examination one year earlier.
Table 1. Clinical findings of patients in two aniridia families.
| Sex | Age (years) |
Vision |
IOP (mmHg) | Aniridia | Cataract | Nystagmus | Other clinical findings | ||
|---|---|---|---|---|---|---|---|---|---|
| OD | OS | ||||||||
| Famliy1 | |||||||||
| II:1 | M | 63 | 0 | 0 | 26/25 | + | + | + | Corneal opacity |
| III:2 | F | 30 | 0.05 | 0.05 | 21/22 | + | + | + | Corneal opacity, microphthalmus |
| Famliy2 | |||||||||
| III:2 | F | 35 | 0.1 | 0.1 | 23/25 | + | + | + | Strabismus, left ectopia lentis |
| IV:1 | M | 12 | 0.4 | 0.3 | 23/24 | + | + | + | Microphthalmus, microcornea, vitreous opacity |
Abbreviations: F, female; IOP, intraocular pressure; M, male; OD, right eye; OS, left eye.
Figure 1.
(a and b) Pedigrees of family 1 and 2 with congenital aniridia. Black symbols indicate affected individuals; white symbols indicate unaffected family members. Diamonds indicate unknown gender. Arrows indicate the probands. Slashed symbols identify deceased subjects. (c and d) Clinical examination details of aniridia patients with PAX6 variants. Family 1: iris photographs show aniridia and corneal opacity both in III2 and II1. Family 2: iris photographs show complete aniridia and left ectopia lentis in III2, with residual iris of left eye in IV1. OD: right eye, OS: left eye.
Molecular genetic analysis
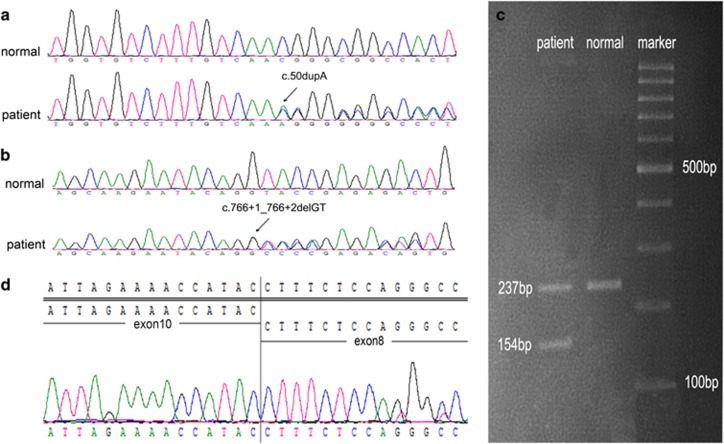
Direct bidirectional sequencing in the proband showed a heterozygous c.50dupA (p.N17Kfs*39) in family 1 and a heterozygous c.765+1_765+2delGT in family 2 (Figures 2a and b), which were also subsequently found in other patients. The variants were not detected in any other unaffected members of the families or in two hundred unrelated controls. They have not been previously reported in the Human Gene Mutation Database, PAX6 Locus Database, dbSNP135, or the Exome Variant Server.
Figure 2.
Genetic characterization of the families with aniridia. (a) A heterozygous duplication (c.50dupA) in PAX6 was found in the proband (III2) and her father (II1) in family 1. (b) A heterozygous splice site deletion (c.765+1_765+2delGT) in PAX6 intron 9 was found in the proband (III2), her father (II1) and her son(IV1) in family 2. The wild-type sequences of unaffected family member are displayed at the top of the figure. The exact variants are indicated by the black arrow and identified. (c) Agarose gel electrophoresis of family 2 showed only one band (237 bp) in unaffected family members, whereas two bands (154 bp and 237 bp) are seen in the proband. (d) Aberrant cDNA sequence revealed skipping of PAX6 exon 9 consequent on the splice site deletion.
Although evaluating the splicing pattern in family 2 by agarose gel electrophoresis, we found an additional amplification product in the proband. Direct sequencing of this PCR product verified the skipping of exon 9, which would lead to a PTC (p.E228Gfs*5; Figures 2c and d).
PAX6 mRNA expression in aniridia patients and unaffected family members
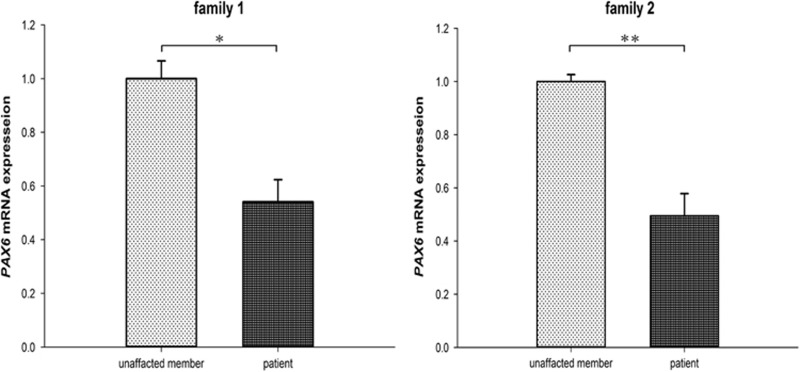
To explore the influence of PTCs on the PAX6 expression in the two families, we reverse transcribed 1 μg of total RNA into cDNA and performed real-time qPCR to compare the expression levels in the aniridia patients and in the unaffected family members. PAX6 mRNA levels were significantly lower in aniridia patients than in unaffected family members, indicating that the duplication and splicing site deletion triggered NMD (P=0.0017 and 0.0006, respectively; Student’s t-test; Figure 3).
Figure 3.
PAX6 mRNA abundance in patients and unaffected family members. The mRNA levels in aniridia patients were significantly lower than in unaffected family members in both families (*P=0.0017, **P=0.0006).
Discussion
Here we report two Chinese families largely suffering from aniridia, cataract, and congenital nystagmus. Moreover, the proband and her father in family 1 showed corneal opacity and microphthalmus. The proband in family 2 was also affected by strabismus and left ectopia lentis, with her son showing a mild progressive cataract and having a high risk of developing angle-closure glaucoma later in his life due to microphthalmus, microcornea, and increased IOP. Our clinical findings present evidence for inter- and intra-familial variable phenotypic manifestations harboring PAX6 variants.12 We even observed slight differences between the two eyes of one patient (proband in family 2), which has seldom been reported.
In family 1, we identified a novel 1-bp duplication (c.50dupA) in PAX6, which was predicted to cause a PTC (p.N17Kfs*39) involving the PD of PAX6. In family 2, we identified a novel 2-bp splice site deletion (c.765+1_765+2delGT) in intron 9, leading to the skipping of exon 9, which was also predicted to generate a PTC (p.E228Gfs*5) in the HD of PAX6. As DNA-binding targets, the PD and HD both played key roles in PAX6 functions.8 Conservation evaluation showed that the changed and deleted residues in the PD and HD were highly conserved across evolutionary timescales, indicating the importance of these domains in ocular development.12, 13 Similar to other frameshift variants at the linker and PST region reported elsewhere in the literature, our patients exhibited the aniridia phenotype, indicating that frameshift variants are overwhelmingly associated with aniridia, regardless of their locations.10, 14, 15 Primignani et al. investigated Italian patients with congenital aniridia and suggested that the variable phenotypes are probably due to different PAX6 variant types. C-terminal extension and splice site variants resulted in phenotypes relatively milder than those consequent of large deletions. In that report, the patient harboring c.765+1G>T (involving the same location as our c.765+1_765+2delGT) variant had no nystagmus, with a visual acuity better than that of patients with other frameshift variants or large deletion.16 However, this phenomenon was not found in family 2, in which the proband had nystagmus, poor visual acuity, and early-onset cataract, showing no obvious differences from the patient in family 1.
According to the available literature, more than three quarters of pathogenic variants predominantly leading to aniridia result in PTCs in the open reading frame.10, 17 Literature based on the haploinsufficiency model suggested that the pathogenic variants resulting in PTCs likely exert pathologic effects through NMD process, during which mRNAs carrying PTCs are rapidly degraded in vivo, a mechanism thought to avoid potentially deleterious consequences of truncated proteins. The NMD surveillance mechanism usually acts if a PTC is located >50–55 nucleotides from the terminal exon-exon junction of PAX6 gene.14, 18, 19 As another possible pathogenic mechanism, frameshift variants located in exon 12 and 13 would escape from NMD and produce truncated proteins, which might exert dominant-negative effect to cause more severe symptoms.20 In our study, the PTCs in the two families with similar clinical manifestations were both located before the last 55 nucleotides of the open reading frame, and the PAX6 mRNA levels of the patients were significantly lower than those of the unaffected members in both families, implying that haploinsufficiency of PAX6 is the major disease mechanism due to NMD. Our study revealed the pathogenicity of two variants and will help us better understand the phenotypes associated with aniridia.
In conclusion, we have identified two novel PAX6 variants in two families with aniridia, cataract, and congenital nystagmus. Our findings expand the variant spectrum of PAX6 gene and provide new evidence for genotype–phenotype correlations, thus facilitating diagnosis and genetic counseling of aniridia.
Acknowledgments
We thank the patients and their families for their cooperation in this project. This work was supported by National Key Basic Research Program of China (2012CB944600) and the National Key Technology R&D Program of China (2012BAI09B05).
Footnotes
The authors declare no conflict of interest.
References
- Song S, Liu Y, Guo S, Zhang L, Zhang X, Wang S et al. A novel PAX6 gene mutation in a Chinese family with aniridia. Mol Vis 2005; 11: 335–337. [PubMed] [Google Scholar]
- Liu Q, Wan W, Liu Y, Liu Y, Hu Z, Guo H et al. A novel PAX6 deletion in a Chinese family with congenital aniridia. Gene 2015; 563(1): 41–44. [DOI] [PubMed] [Google Scholar]
- Nelson LB, Spaeth GL, Nowinski TS, Margo CE, Jackson L. Aniridia. A review. Surv Ophthalmol 1984; 28(6): 621–642. [DOI] [PubMed] [Google Scholar]
- Hanson IM. PAX6 and congenital eye malformations. Pediatr Res 2003; 54(6): 791–796. [DOI] [PubMed] [Google Scholar]
- Brauner SC, Walton DS, Chen TC. Aniridia. Int Ophthalmol Clin 2008; 48(2): 79–85. [DOI] [PubMed] [Google Scholar]
- Fischbach BV, Trout KL, Lewis J, Luis CA, Sika M. WAGR syndrome: a clinical review of 54 cases. Pediatrics 2005; 116(4): 984–988. [DOI] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet 2002; 18(1): 41–47. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. NAT GENET 1992; 2(3): 232–239. [DOI] [PubMed] [Google Scholar]
- Brown A, McKie M, van Heyningen V, Prosser J. The human PAX6 mutation database. Nucleic Acids Res 1998; 26(1): 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulaki I, White IM, Hanson IM. PAX6 mutations: genotype-phenotype correlations. BMC Genet 2005; 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronskov K, Rosenberg T, Sand A, Brondum-Nielsen K. Mutational analysis of PAX6: 16 novel mutations including 5 missense mutations with a mild aniridia phenotype. Eur J Hum Genet 1999; 7(3): 274–286. [DOI] [PubMed] [Google Scholar]
- Hingorani M, Williamson KA, Moore AT, van Heyningen V. Detailed ophthalmologic evaluation of 43 individuals with PAX6 mutations. Invest Ophthalmol Vis Sci 2009; 50(6): 2581–2590. [DOI] [PubMed] [Google Scholar]
- Chen JH, Lin W, Sun G, Huang C, Huang Y, Chen H et al. A novel PAX6 deletion in a Chinese family with congenital aniridia. Mol Vis 2012; 18(1): 989–995. [PMC free article] [PubMed] [Google Scholar]
- Dubey SK, Mahalaxmi N, Vijayalakshmi P, Sundaresan P. Mutational analysis and genotype-phenotype correlations in southern Indian patients with sporadic and familial aniridia. Mol Vis 2015; 21: 88–97. [PMC free article] [PubMed] [Google Scholar]
- Hingorani M, Hanson I, van Heyningen V. Aniridia. Eur J Hum Genet 2012; 20(10): 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primignani P, Allegrini D, Manfredini E, Romitti L, Mauri L, Patrosso MC et al. Screening of PAX6 gene in Italian congenital aniridia patients revealed four novel mutations. Ophthalmic Genet 2016; 37(3): 307–313. [DOI] [PubMed] [Google Scholar]
- Neethirajan G, Solomon A, Krishnadas SR, Vijayalakshmi P, Sundaresan P. Genotype/phenotype association in Indian congenital aniridia. Indian J Pediatr 2009; 76(5): 513–517. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 1999; 96(3): 307–310. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004; 5(2): 89–99. [DOI] [PubMed] [Google Scholar]
- Singh S, Tang HK, Lee JY, Saunders GF. Truncation mutations in the transactivation region of PAX6 result in dominant-negative mutants. J Biol Chem 1998; 273(34): 21531–21541. [DOI] [PubMed] [Google Scholar]