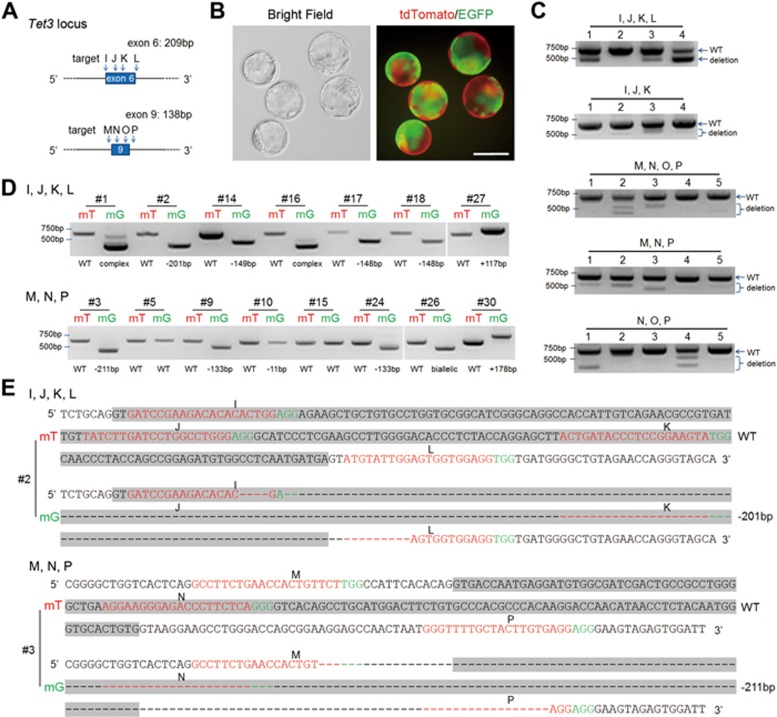
Figure 2.
Gene editing using multiple sgRNAs efficiently produced chimeras carrying large deletions or insertions. (A) Schematic of the Cas9/sgRNA-targeting sites in exons 6 and 9 of Tet3. (B) Representative mT+/mG+ blastocysts generated by combinatorial MNP sgRNAs targeting Tet3 in one blastomere of two-cell stage embryos through 2CC. Scale bar, 100 μm. (C) Single blastocyst PCR genotyping of blastocysts derived from different combinations of sgRNAs via 2CC method. (D) Representative PCR results of mT+/mG+ chimeric mice generated using the IJKL or MNP sgRNA combinations. Most mT+/mG+ mice carried large deletions or insertions in mG+ cells. “Complex” denotes multiple mutations (three or more). “Biallelic” here denotes one apparent PCR band from mouse #26, which upon sequencing showed two distinct mutations. (E) Representative sequencing results of mT+/mG+ chimeric mice generated using the IJKL or MNP sgRNA combinations. The sgRNA-targeting sequence is labeled in red, and the protospacer-adjacent motif (PAM) sequence is labeled in green. Sequences of exons 6 and 9 are highlighted in gray.