Abstract
Bacteriophages encode anti-CRISPR suppressors to counteract the CRISPR/Cas immunity of their bacterial hosts, thus facilitating their survival and replication. Previous studies have shown that two phage-encoded anti-CRISPR proteins, AcrF1 and AcrF2, suppress the type I-F CRISPR/Cas system of Pseudomonas aeruginosa by preventing target DNA recognition by the Csy surveillance complex, but the precise underlying mechanism was unknown. Here we present the structure of AcrF1/2 bound to the Csy complex determined by cryo-EM single-particle reconstruction. By structural analysis, we found that AcrF1 inhibits target DNA recognition of the Csy complex by interfering with base pairing between the DNA target strand and crRNA spacer. In addition, multiple copies of AcrF1 bind to the Csy complex with different modes when working individually or cooperating with AcrF2, which might exclude target DNA binding through different mechanisms. Together with previous reports, we provide a comprehensive working scenario for the two anti-CRISPR suppressors, AcrF1 and AcrF2, which silence CRISPR/Cas immunity by targeting the Csy surveillance complex.
Keywords: CRISPR/Cas system, anti-CRISPR proteins, phage, immune evasion, cryo-EM
Introduction
Bacteria and archaea have evolved numerous mechanisms to fight against phage infection (antiviral systems)1. One of the most typical representatives is the CRISPR/Cas system that relies on CRISPR (clustered regularly interspaced short palindromic repeats) loci and a diverse cassette of CRISPR-associated (Cas) genes2. It has been estimated that more than 95% archaea and 48% eubacteria possess one or more types of CRISPR/Cas systems3. However, to establish infection and proliferate in bacterial hosts, phages have also evolved sophisticated strategies to evade the CRISPR/Cas immunity. Recently, the newly identified anti-CRISPR proteins, which can suppress host bacterial CRISPR/Cas systems and facilitate phage survival, attracted tremendous attention4,5,6,7. The interactions of CRISPR/Cas systems with anti-CRISPR suppressors are the results of virus and host co-evolution, which provides new insights into the development and functions of CRISPR/Cas systems.
The CRISPR/Cas immunity involves three stages, including spacer acquisition, CRISPR RNA (crRNA) production and interference8. Initially, foreign nucleic acid fragments are inserted as spacers into the CRISPR locus in bacterial genomes, which serve as a library for invading nucleic acid9,10,11. The CRISPR array, containing multiple spacers and CRISPR repeats, is then transcribed into pre-crRNA that would be further processed into mature crRNA with only one spacer sequence and repeat12,13. Following that, Cas proteins and crRNA assemble into a ribonucleoprotein complex to detect the complementary target nucleic acid based on the foreign spacer sequence and further induce target degradation14,15,16.
According to the unified classification of CRISPR/Cas systems, all type I systems utilize a multi-subunit surveillance complex to recognize the complementary invading nucleic acid, and then a Cas endonuclease is recruited to degrade the target, typically Cas3 protein family members14,15,16,17,18. All these CRISPR/Cas effectors could potentially be intercepted by phage-encoded anti-CRISPR proteins to inactivate the CRISPR/Cas immunity. Previous studies have identified a set of anti-CRISPR (Acr) proteins targeting type I-F CRISPR/Cas system of Pseudomonas aeruginosa, which counteract the CRISPR/Cas immunity at different stages4,19. Among them, AcrF3 directly interacts with Cas3 endonuclease to lock it in ADP-binding conformation, thus inhibiting its enzymatic activity20,21. AcrF1 and AcrF2 bind to the surveillance complex, Csy complex in this system, preventing target DNA recognition19. Biochemical analysis has located their binding sites at the Csy3 backbone and Csy1-Csy2 tail of the Csy complex, respectively19,22. However, the structural basis of AcrF1 and AcrF2 inhibiting DNA recognition by Csy complex remains obscure.
To elucidate the mechanism of AcrF1- and AcrF2-mediated silencing of type I-F CRISPR/Cas system, we determined the structures of AcrF1 and AcrF2 bound to Csy complex (AcrF1/2-Csy complex) by cryo-electron microscopy (cryo-EM) at near atomic resolution. We precisely located AcrF1 and AcrF2 in this complex and found multiple modes of these two anti-CRISPR suppressors binding to Csy complex, demonstrating their alternate mechanisms working individually or synergistically. During the preparation of this manuscript, a similar complex structure was published, which reported only one binding mode23. Our results provide a comprehensive working scenario of these two anti-CRISPR effectors, which would intensify our understanding of the molecular mechanisms of anti-CRISPR proteins to silence the CRISPR/Cas immunity by targeting surveillance complexes.
Results
Cryo-EM reconstruction
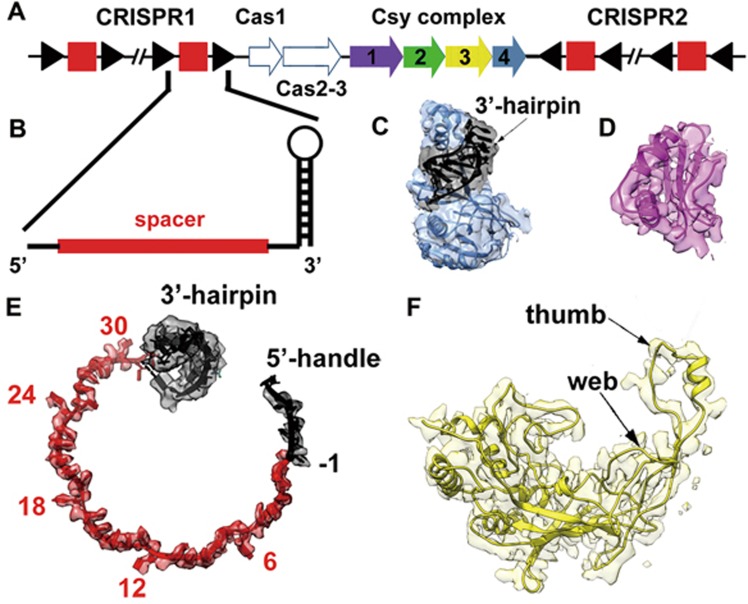
Initially, we prepared AcrF1 and AcrF2 bound to Csy complex with a 32-nt spacer crRNA (AcrF1/2-Csy32nt complex), which represents the naturally occurring Csy complex in P. aeruginosa (Figure 1A and 1B; Supplementary information, Figure S1A). The backbone part of this complex, containing six copies of Csy3 and two copies of AcrF1, was well reconstructed to an overall resolution of 3.8 Å (Supplementary information, Figure S2C). However, the Csy4 head and Csy1-Csy2 tail were poorly resolved with large part of omit density, reflecting the intrinsic flexibility of these two parts in this complex (Supplementary information, Figures S2B, S3, S6A and S6B). Besides, the orientation preference problem also limited the reconstruction result, making it difficult to build atomic models (Supplementary information, Figure S2A, S2B and S2D).
Figure 1.
EM density of the modeled components in the reconstructed complex. (A) Schematic representation of the CRISPR/Cas gene clusters of P. aeruginosa. The CRISPR repeats and spacers are represented as black triangles and red squares, respectively. The Cas2 protein in this system is fused as a domain in the Cas3 protein. The four Csy proteins consisting of the Csy complex (Csy1-4) are consecutively arranged in the genome and colored in purple, green, yellow and slate, respectively. (B) Close view of the schematic model of a mature crRNA with the spacer colored in red and repeat sequence in black. (C) The density of Csy4-crRNA hairpin head and the fitted atomic model. The Csy4 protein is represented by slate ribbons and the crRNA 3′-hairpin is highlighted in black. (D) Representative density of AcrF1 and the refined atomic model. The model is shown as ribbons and the density map shows clear side chain features. (E) The de novo built model of crRNA 5′-handle and spacer chain is shown as ribbons and colored in the same manner as in B. The rigidly fitted 3′-hairpin is also shown to give an overall structure of the crRNA molecule. The kink nucleotides are labeled by the positions in spacer sequence. The density map shows clear side chains of each nucleotide. (F) Representative density map and model of Csy3 subunit. The thumb and web domains are labeled aside.
The closest relative of type I-F CRISPR/Cas system is the intensively studied type I-E system, of which the crRNA encapsidating mechanism of the surveillance complex (Cascade complex) has been well defined18. With this reference, we reduced the crRNA spacer to 20 nucleotides, which resulted in a smaller complex with four copies of Csy3 and a single copy of AcrF1 (AcrF1/2-Csy20nt complex) (Supplementary information, Figures S1B, S4B and S5). The smaller complex behaved better in orientation distribution, and the reconstructed map was improved, with which we can produce a full component reconstruction with an overall resolution of 5.3 Å and focused-refine the backbone to a resolution of 4.2 Å (Supplementary information, Figures S4A-S4D, S6C and S6D). The improved reconstruction helped to recognize the topology of Csy3 protein and better resolved the Csy4 head and Csy1-Csy2 tail, which allowed us to build an atomic model for the Csy3-crRNA-AcrF1 subcomplex and rigidly fit Csy4 and crRNA hairpin (PDB: 4AL5) unambiguously into the density (Figure 1C-1F). The reported AcrF1 atomic model (PDB: 2LW5) was perfectly fitted into the corresponding density map, which helped to locate each protein component (Figure 1D). The density of crRNA side chains was clearly recognizable and facilitated our sequence registration (Figure 1E). The model of Csy3 was de novo built based on the two backbone maps with 3.8 and 4.2 Å resolutions, respectively. Initially the topology was identified and the main chain was traced with poly-alanine. The bulky amino-acid side chains were identified to help update the sequence register and then a near-complete Csy3 atomic model was built (Figure 1F). The final atomic model contains Csy3-Csy4-crRNA-AcrF1 components, whereas the Csy1-Csy2-AcrF2 subcomplex could only be located but not modeled.
Structural features of Csy complex and comparison with Cascade complex
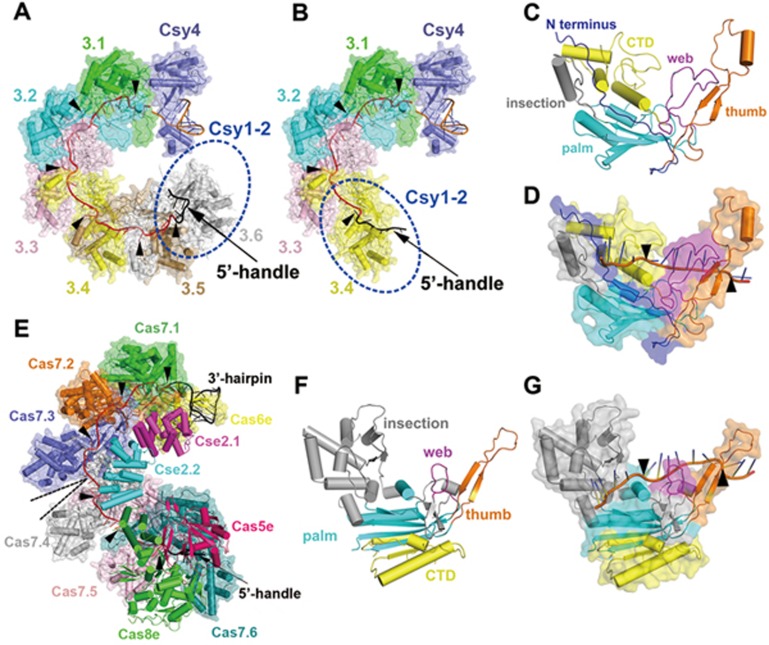
The overall structure of Csy32nt complex showed a flat spiral architecture, with the Csy4 head and Csy1-Csy2 tail rolling to proximity and forming a nearly closed ring structure (Figure 2A). The backbone contains six copies of Csy3 molecules, which are well ordered following helical symmetry. Along the helical track, the crRNA spacer was accommodated in a groove formed by the Csy3 backbone (Figure 2A). Six nucleotides of RNA segment were involved in interacting with each copy of Csy3 subunit, and a kink was observed among every 6 nucleotides (Figure 2A, 2B and 2D). This feature highly resembles the RNA presenting modes of the Cascade complex in type I-E system (Figure 2E and 2G) and Cmr complex in type III-B system, indicating that it might be a universal mechanism of crRNA encapsidation in all class I CRISPR/Cas systems16,17,18,24.
Figure 2.
Overall structure of Csy surveillance complex and comparison with Cascade complex. (A) Cartoon and surface representation of the overall structure of Csy3-Csy4-crRNA subcomplex (32-nt spacer). The unmodeled Csy1-Csy2 tail is represented by a blue dash line ellipsoid. The crRNA spacer and 5′-handle are colored in red and black, respectively. Six copies of Csy3 subunit are colored by chains and the kink nucleotides are labeled by black triangles along the crRNA chain. (B) The modified Csy3-Csy4-crRNA subcomplex (20-nt spacer) atomic model shown in the same style as in A. (C) Structure of Csy3 subunit in cartoon representation. The N-terminus, palm, thumb, web and C-terminal domains are shown in different colors. The web loop is highlighted in magenta. (D) Surface and cartoon representation of Csy3 subunit showing the encapsidated crRNA segment. The kink nucleotides are highlighted by black triangles. (E) Overall structure of Cascade complex shown in the same style of Csy complex as in A. The loosely arranged Cas7 backbone is shown as surface and cartoon with the space between adjacent Cas7 subunits highlighted by black dash lines. (F) The overall structure of Cas7 subunit shown in the same manner as Csy3 in C. (G) A Cas7 subunit interacting with crRNA fragment shown in the same fashion as Csy3 in D.
In addition to these common structural features, substantial differences were also observed in the Csy32nt complex compared to the Cascade complex. The adjacent Csy3 molecules are more tightly assembled than Cas7 subunits in Cascade complex, making the helical rise of Csy complex smaller than that of Cascade complex (Figure 2A and 2E). As a result, the crRNA spacer segment encapsidated in Csy complex is placed in a relatively narrower groove in contrast to the more exposed crRNA presenting mode of Cascade complex (Figure 2A and 2E). More strikingly, the six copies of Csy3 molecules are assembled in the same orientation along the helical track (Figure 2A), whereas the Cas7 molecule adjacent to Cas8e-Cas5e tail (Cas7.6) rotated ∼160° relative to other five copies due to interactions with Cas5 subunit in the Cascade complex17,18,25. This observation indicates that the interactions between Csy2 (Cas5f) and Csy3.6 may follow a substantially different mechanism, thus creating a different double-strand DNA (dsDNA) binding site at the tail region. This highly ordered helical symmetry also allowed us to superimpose the well-resolved AcrF1/2-Csy32nt complex backbone with the AcrF1/2-Csy20nt complex with all components resolved, thus reconstituting a full-component AcrF1/2-Csy32nt complex (Figure 3A; Supplementary information, Figure S7A and S7B).
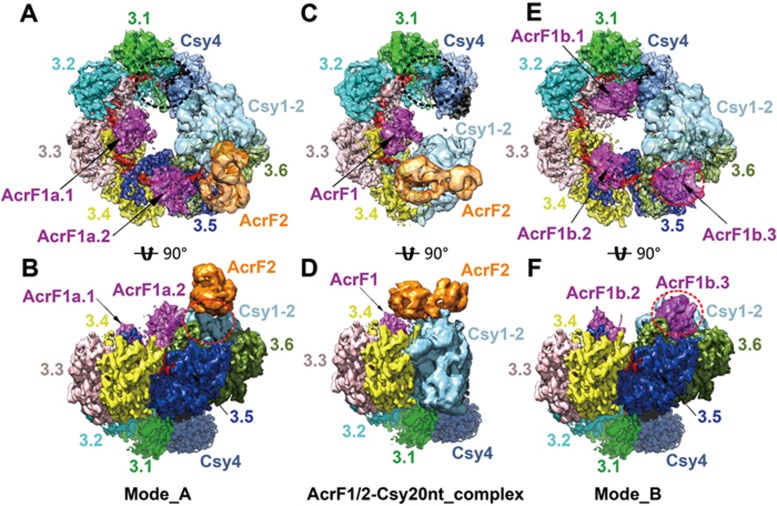
Figure 3.
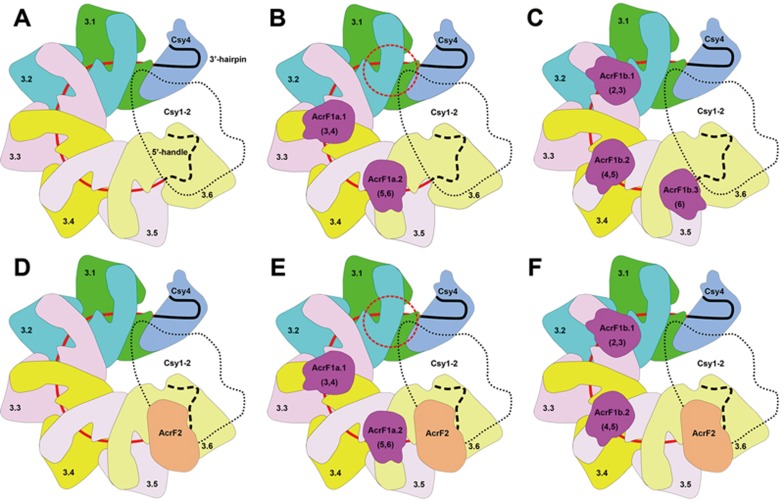
Cryo-EM reconstructed map models of AcrF1/2-Csy complexes with different AcrF1/2 binding modes. (A, B) Top and side views of full-component AcrF1/2-Csy32nt complex with two copies of AcrF1 and one copy of AcrF2 binding to Csy32nt complex simultaneously (mode A). The Csy3-crRNA-AcrF1 backbone and Csy4 head are shown with both ribbon models and EM density surfaces. Six copies of Csy3 subunits are colored by chains. The AcrF1 and AcrF2 molecules are colored in magenta and orange, respectively. The Csy1-Csy2-AcrF2 tail is shown with only density map without models. (C, D) The modified AcrF1/2-Csy20nt complex is shown in top and side views in the same style as A and B. (E, F) Csy32nt complex with three copies of AcrF1 binding (mode B) is shown in the same views as mode A in A and B. The free AcrF1 binding sites at Csy3.(1-2) site in A and C are indicated by black dash line ellipsoids, which are partially interfered by the Csy4 head. The positions of AcrF1b.3 in E and F are highlighted by red dash line ellipsoids and the corresponding site in B is indicated similarly to show the spatial clash with AcrF2.
The Csy3 density was well resolved and an atomic model was successfully built. The overall structure of Csy3 molecule follows the general domain architecture of Cas7 family members, composed of a palm domain and C-terminal domain in the main body, a thumb loop reaching out and a web domain loop interspaced in between (Figure 2C). The palm domain contains a conserved RNA recognition motif, which is the major part responsible for crRNA binding. The thumb loop intersects into the space between the main body and thumb loop of another adjacent Csy3 subunit, thus allowing Csy3 molecules to self-assemble following helical symmetry (Figure 2A and 2B; Supplementary information, Figure S7C), which is a general mechanism of assembly for all Cas7 family members and shapes the backbone of surveillance complexes16. The crRNA is accommodated in the groove formed by Csy3 main body and thumb loop, in which every Csy3 subunit interacts with six nucleotides and the thumb loop induces a kink among every 6-nt crRNA segment in a similar manner to Cas7 in the Cascade complex (Figure 2A, 2D, 2E and 2G). However, the interspaced web loop of Csy3 is much longer than that of Cas7 and is directly involved in the interaction with crRNA spacer (Figure 2C, 2D, 2F and 2G). This makes the crRNA binding groove narrower compared with the highly-exposed crRNA cradle in the Cascade complex, which might lead to a different target DNA recognition mechanism of the Csy complex in type I-F CRISPR/Cas system.
Binding modes of AcrF1 and AcrF2
In the cryo-EM reconstruction map, we clearly recognized the density of AcrF1 and AcrF2 molecules in the complex, which facilitated our structural analysis to explain the working mechanisms of these two anti-CRISPR suppressors. In the full-component AcrF1/2-Csy32nt complex, one copy of AcrF2 was observed to bind to the Csy1-Csy2 tail and two copies of AcrF1 were found to bind to the Csy3 backbone inside the helical ring (Figure 3A and 3B), which is in good accordance with previous biochemical evidence and a similar cryo-EM structure reported recently19,23.
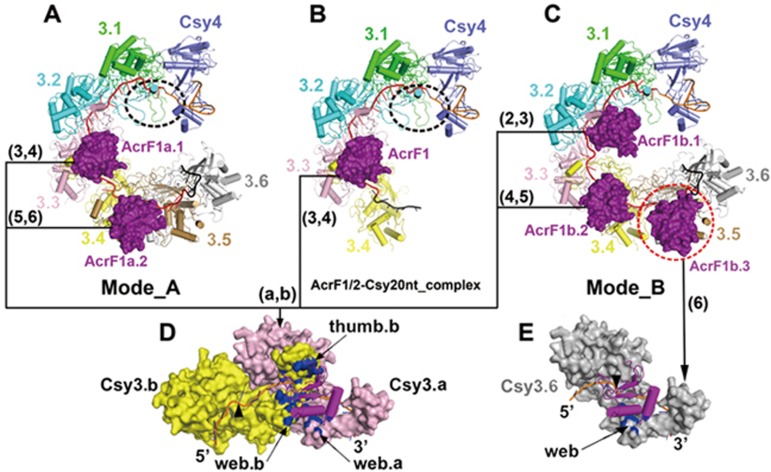
However, in addition to this binding mode (mode A), we also observed another AcrF1-alone binding mode (mode B; Figure 3E and 3F). During image processing, we noticed that the 3D classification of original AcrF1/2-Csy32nt complex particles resulted in two main types of particles, with two or three copies of AcrF1 binding, respectively (Supplementary information, Figure S3). Comparing these two binding modes, we found that the AcrF1 binding sites in the two structures were not overlapping but interlaced to each other (Figure 3A, 3B, 3E and 3F). This phenomenon was quite conceivable as all six copies of Csy3 subunits assembled in the same orientation following ordered helical symmetry, thus five equivalent interfaces were created between every two adjacent Csy3 subunits, which is right the binding site of AcrF1. However, it is impossible for all five positions to be occupied by AcrF1 simultaneously due to steric hindrance between two AcrF1 molecules at adjacent binding sites (Figure 3A-3F).
In binding mode A, which is consistent with the observation by Wiedenheft's group23, two AcrF1 (AcrF1a.1 and AcrF1a.2) molecules bind to the interface formed by Csy3.(3-4) and Csy3.(5-6), respectively, but leaving the Csy3.(1-2) interface free, which might be interfered by spatial clash rendered by the Csy4 head (Figures 3A, 3C, 4A and 4B; Supplementary information, Figure S8E and S8F). Thus only two copies of AcrF1 molecule could bind to Csy32nt complex in this mode. In the alternative binding mode (mode B), however, two equivalent interfaces formed by Csy3.(2-3) and Csy3.(4-5) are occupied by two AcrF1 (AcrF1b.1 and AcrF1b.2) molecules and the third AcrF1 (AcrF1b.3) binds to a single Csy3.6 subunit, which is in close proximity to the Csy1-Csy2 tail region (Figure 3E and 3F). Upon the AcrF1b.3 binding, one end of the AcrF2 binding site on Csy1-Csy2 tail was blocked, thus generating steric hindrance for the binding of AcrF2 molecule (Figure 3A, 3B, 3E and 3F). In this situation, only AcrF1 molecules could bind to the Csy complex, which represents the scenario of AcrF1 working alone. In the smaller AcrF1/2-Csy20nt complex, only one AcrF1 binds at the Csy3.(3-4) interface and one AcrF2 binds at the Csy1-Csy2 tail region, which is consistent with the binding mode A of AcrF1/2-Csy32nt complex.
Figure 4.
Atomic models of Csy3-Csy4-crRNA-AcrF1 subcomplexes corresponding to different AcrF1/2 binding modes. The Csy complex components are shown as cartoon and colored by chains. AcrF1 molecules are highlighted in magenta and represented by surface models. The Csy3 subunits interacting with each copy of AcrF1 are labeled aside the corresponding AcrF1 molecules. (A, C) Model of AcrF1 binding to Csy32nt complex in modes A and B, respectively. (B) Model of AcrF1 binding to Csy20nt complex. The free AcrF1 binding sites at Csy3.(1-2) site in A and B are indicated by black dash line ellipsoids, which are partially blocked by Csy4 head. (D, E) Close view of AcrF1 interacting with Csy3 subunits. The interaction interfaces on Csy3 subunits are highlighted by blue color. The kink nucleotides in crRNA segment are labeled by black triangles. D shows the general interaction mode that involves two copies of Csy3 molecules and E presents the third copy of AcrF1 in binding mode B (AcrF1b.3) interacting with a single Csy3 subunit (Csy3.6).
As previous studies have demonstrated that either AcrF1 or AcrF2 is sufficient to silence the Csy complex-mediated CRISPR/Cas immunity4,19, the two binding modes of AcrF1/2 to Csy complex exactly reveal how they work synergistically or individually. AcrF2 has only one binding site at the Csy1-Csy2 tail region, and thus only one binding mode to prevent the DNA target recognition by the Csy complex, which shows no difference when working alone or cooperating with AcrF1 (Figure 3A-3D). However, AcrF1 could adopt at least two binding modes to inhibit Csy complex function when working alone, which involve at most two or three copies of AcrF1 at the Csy3.[(3-4),(5-6)] or Csy3.[(2-3),(4-5),(6)] interaction sites, respectively. In addition, when synergistically working together with AcrF2, no more than two copies of AcrF1 could participate in the interaction (Figure 3A, 3B, 3E and 3F).
Molecular mechanisms of inhibiting CRISPR/Cas immunity by AcrF proteins
Based on the atomic model of Csy3-crRNA-AcrF1 subcomplex, we further analyzed the detailed interactions between Csy complex and AcrF1 suppressors in different binding modes. As the six Csy3 subunits are arranged in the same orientation, the four equivalent binding sites, Csy3.[(2-3),(3-4),(4-5),(5-6)], all involve two adjacent Csy3 molecules to create a three-component interface, including the web loop (web.b) and thumb loop (thumb.b) from one Csy3 subunit and the web loop from the other (web.a) (Figure 4A-4D; Supplementary information, Figure S8A-S8C). However, for the AcrF1b.3 in binding mode B, only the web loop of Csy3.6 is responsible for the interaction (Figure 4C and 4E; Supplementary information, Figure S8D). It is not clear whether AcrF1b.3 interacts with the Csy1-Csy2 tail, as this region was poorly resolved in our reconstruction. Considering their proximity in location, Csy1-Csy2 tail might help to stabilize the binding of AcrF1b.3.
For the Cascade complex in type I-E CRISPR/Cas system, each Cas7 subunit accommodates six nucleotides of crRNA spacer with one displaced kink behind the thumb domain17,18. The other five nucleotides are readily accessible for target DNA base-pairing, leading to periodic five base pairs interspaced by one-nucleotide kinks26. This is a general mechanism for target DNA recognition shared by all surveillance complexes in class I CRISPR/Cas systems16. Therefore, binding of AcrF1 to the thumb of Csy3 subunit would prevent the target DNA base-pairing with the crRNA spacer (Figure 4D).
From Wiedenheft group's report, AcrF2 is a dsDNA mimic to compete the binding site of the target DNA duplex on Csy1-Csy2 tail23. As AcrF1b.3 and AcrF2 are mutually exclusive in binding the Csy complex in binding mode B, we deduce that AcrF1b.3 could also prevent the dsDNA binding by potential steric hindrance in a similar manner to interfering with AcrF2 binding (Figure 3E, 3F, 4C and 4E). Thus, AcrF1 can utilize different mechanisms to block target DNA recognition of the Csy complex when adopting different binding modes (Figure 5A-5F).
Figure 5.
Schematic models of AcrF1/2 interacting with Csy complex in different binding modes. Each Csy3 subunit of Csy complex is indicated by a unique color and the Csy1-Csy2 tail is shown as a single part in the model. The crRNA spacer is colored in red and the 5′-handle and 3′-hairpin are colored in black. AcrF1 and AcrF2 molecules are colored in magenta and orange, respectively. The crRNA 5′-handle is underneath the Csy1-Csy2 tail, which is invisible in the top view, thus represented by dash lines. The Csy1-Csy2 tail is not colored and set as transparent to show the Csy4 head and Csy3.1 subunit underneath. (A) Schematic structure of Csy32nt complex. (B, C) Model of AcrF1 alone binding to Csy complex in modes A and B, respectively. The potential AcrF1 binding site interfered by Csy4 head is indicated by a red dash line ellipsoid. (D) Model of AcrF2 binding to Csy complex alone. (E, F) Model of AcrF1 and AcrF2 binding to Csy complex simultaneously where AcrF1 adopts different binding modes. E and F represent the binding mode A and B, respectively.
Discussion
Anti-CRISPR protein suppressors represent a newly-established research direction in the CRISPR/Cas field, which is the result of co-evolution between prokaryotic organisms and their parasitic phages7. Virtually, the interactions between anti-CRISPR proteins and CRISPR/Cas systems reflect a sophisticated strategy that viruses adopt to evade host immunity. It is quite conceivable that many unknown anti-CRISPR suppressors exist and are yet to be identified.
Anti-CRISPR proteins targeting the P. aeruginosa type I-F CRISPR/Cas system have been reported to function at different stages, among which AcrF3 targets Cas3 endonuclease to inhibit its DNA cleavage activity whereas AcrF1 and AcrF2 interfere with the DNA target recognition by targeting the Csy surveillance complex19. The structural basis of inactivating Cas3 endonuclease by AcrF3 has been well characterized as locking Cas3 in an ADP-binding conformation that is incapable of cleaving DNA target20,21. However, the molecular mechanisms of preventing the Csy complex from recognizing target DNA by AcrF1 and AcrF2 remain obscure.
Here, our findings provide comprehensive insights into the multiple working modes of AcrF1 and AcrF2 to silence type I-F CRISPR/Cas immunity. There is only one binding site for AcrF2 on the Csy complex, and thus the inhibition mechanism is simply preventing dsDNA binding. By contrast, the binding of AcrF1 to the Csy complex is multivalent, suggesting sophisticated working mechanisms when adopting different binding modes. Though AcrF1 and AcrF2 bind at different sites on the Csy complex, the binding mode of AcrF1 could be affected by AcrF2, as one of the AcrF1 binding sites (AcrF1b.3 binding site) is close to the AcrF2 binding site. Due to the spatial incompatibility of these two molecules at this region, their binding is mutually exclusive, thereby shaping the diversity of AcrF1 binding modes. Since AcrF2 mimic the dsDNA target and AcrF1b.3 excludes the binding of AcrF2, AcrF1 binding at this site might also prevent the binding of dsDNA target, which is different from AcrF1 molecules at other binding sites that prevent single-strand DNA base-pairing with the crRNA spacer. For the interactions of AcrF1 with the Csy complex, the Csy3 web loop plays a major role. As the web loop region of Csy3 shows significant difference from that of Cas7 in the Cascade complex, it might explain why AcrF1 specifically targets only the type I-F CRISPR/Cas system in P. aeruginosa4,19.
It has been demonstrated that either AcrF1 or AcrF2 alone is sufficient to block the activity of the Csy complex4,19. Based on the structural studies, a single copy of AcrF2 is capable of silencing the CRISPR/Cas immunity but it is not clear how many copies of AcrF1 are enough to suppress DNA target recognition by the Csy complex. According to the above analysis, one copy of AcrF1 should be effective to exclude the binding of DNA duplex once placed at the AcrF1b.3 binding site. However, if binding to other sites, one copy of AcrF1 might not be sufficient to block the DNA binding, as a large proportion of the spacer sequence could still be paired with the DNA target. More efforts are required to address this issue in the future.
Uncovering the molecular mechanisms of silencing CRISPR/Cas immunity by various anti-CRISPR proteins is of significant theoretical importance to improve our understanding of functionality of CRISPR/Cas system and to guide their engineering for application. To date, we have attempted to apply phages to deal with many pathogenic bacteria, however, the bacterial CRISPR/Cas-related immune systems can affect the activity of phages27,28,29,30. Developing novel anti-bacterial therapeutics based on anti-CRISPR suppressors is a very promising strategy, especially for dealing with the re-emergent drug-resistant bacterial infections.
Materials and Methods
Protein expression and purification
The full-length genes encoding AcrF1 (GenBank: JX434032.1), AcrF2 (GenBank: NC_005178) and P. aeruginosa Csy proteins (Csy1-4, GenBank: CP000438.1) were synthesized and codon optimized for expression in Escherichia coli. The coding sequences of AcrF1 and AcrF2 with an N-terminal 6× His tag followed by tobacco etch virus (TEV) protease cleavage site were inserted individually into the expression vector pET21a. The Csy genes were sequentially subcloned into a modified polycistronic expression vector based on pET21a, and only the Csy4 coding sequence contains a 6× His affinity tag and a TEV protease cleavage site at the N-terminus. crRNA-encoding genes with 32- or 20-nt spacer were synthesized and ligated into pET28a vector backbone driven by a T7 promoter. Expression vectors of Csy proteins and crRNA were co-transformed into E. coli BL21 (DE3) strain for Csy complex expression.
For expression of all individual proteins or Csy complex, the cells were induced with 0.5 mM isopropyl-β-𝒟-thiogalactopyranoside (IPTG) at an OD600 nm of 0.6 and grown at 16 °C for an additional 15 h. Cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl) and homogenized with a low-temperature ultrahigh-pressure cell disrupter (JNBIO, China). Proteins were initially purified by Ni-NTA affinity chromatography (GE Healthcare). For AcrF1 and AcrF2, the eluted product was pooled and buffer exchanged into 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and then digested with TEV protease at 18 °C for 12 h. The cleaved protein samples were further purified by size-exclusion chromatography with a Superdex 200 10/600 pg (GE Healthcare) column. For Csy complex, the eluent was concentrated and dialyzed at 4 °C overnight against ion-exchange buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl) for further purification with a Resource Q column (GE Healthcare). Then the complete complex was cleaved by TEV protease and purified by another round of size-exclusion chromatography.
To prepare the AcrF1/2-Csy complex, AcrF1 and AcrF2 proteins were simultaneously mixed with Csy complex at a molar ratio of 4:1 and 1.5:1, respectively, and incubated at 18 °C for 2 h prior to purification with a Superose 6 10/300 GL size-exclusion column (GE Healthcare). The resulting complex reached a purity of ∼95% as shown by SDS-PAGE and protein concentrations were quantified by measuring A280.
Cryo-EM sample preparation and data collection
Purified AcrF1/2-Csy complex sample (3 μl) with a concentration of 0.4 mg/ml or 0.7 mg/ml, for samples with 32- or 20-nt spacers, respectively, was placed on a glow-discharged holy carbon grid (Quantifoil 1.2/1.3 holy carbon, 300 mesh). After 2.5 s blotting with filter paper, the grid was flash plunged in liquid ethane using a Vitrobot Mark IV (FEI Company) at 4 °C and 100% humidity. Cryo-EM single-particle data collection was performed using a 300 kV Titan Krios microscope equipped with a K2 summit camera. Images were recorded with super-resolution mode and dose-fractionized into 38-frame movies.
For AcrF1/2-Csy32nt complex, each image was exposed for 7.6 s at a calibrated magnification of 38 461 and an electron dose rate of ∼4.73 e−/Å2/s, resulting in a total dose of ∼36 e−/Å2. The images were binned before data processing, yielding a final pixel size of 1.3 Å. For AcrF1/2-Csy20nt complex, images were taken with a different microscope with similar setup, which shows slight difference in magnification effect. Each exposure lasted 11.4 s at a calibrated magnification of 38 168 and an electron dose rate of ∼5.17 e−/Å2/s, giving an accumulative dose of ∼59 e−/Å2. The images were binned to a final pixel size of 1.31 Å before reconstruction.
Image processing
The distortion and beam induced motion of each image stack were corrected by MAG_DISTORTION_CORR_v8.18.1531 and MOTIONCORR_v2.132, respectively. The full stack sum was calculated for subsequent processing and the parameters of the contrast transfer function (CTF) were determined by CTFFIND33. A subset of protein particles were semi-automatically boxed using the program e2boxer.py in EMAN234 software package and two dimensional (2D) classified using RELION-1.435. Four distinguished class average images were selected as the reference for automatic particle picking of the whole data set by RELION-2.036.
For AcrF1/2-Csy32nt complex, a total number of ∼530 000 initial particles were picked in 3 235 micrographs and 2 rounds of reference-free 2D classification were performed to clean up the data set, which yielded a subset of ∼260 000 particles in the good classes showing clear secondary structure features (Supplementary information, Figure S2A and S2B). The selected particles were subjected to global 3D classification followed by local search. All particles converged into two main types of reconstructions in which AcrF1 molecules bind at different sites, designated as binding modes A and B, respectively (Supplementary information, Figure S3). In all reconstructed maps, both the Csy4 head and Csy1-Csy2 tail were poorly resolved, indicating intrinsic flexibility of these two regions. In the reconstructed map of a main class in binding mode B, both the head and tail regions showed better densities compared with other classes. The particles in this class were subjected to 3D refinement and yielded a 4.7 Å resolution reconstruction (Supplementary information, Figure S3). As the head and tail regions showed different extent of missing density, a soft mask covering the rigid backbone was applied to the 3 main classes in mode A for focused refinement, resulting in a 3.95 Å resolution map (Supplementary information, Figure S3). At the final stage, dose reduction was performed to further improve the resolution. Briefly, a new stack containing frame 3-23 in each 38-frame full-dose stack was extracted and re-aligned using MOTIONCORR_v2.132, which resulted in a total dose of ∼20 e−/Å2. The low-dose data set was applied for final refinement and the resolution was improved to 3.77 Å with the 0.143 Fourier shell correlation cutoff (Supplementary information, Figure S2C).
For AcrF1/2-Csy20nt complex data set, ∼390 000 particles were automatically picked from 599 images and 2D classified for 3 rounds to remove false positive and bad particles, which resulted in a clean subset of ∼180 000 particles. A single round of 3D classification was conducted to further distinguish the structural heterogeneity, which started with vigorous global search followed by restricted local search. Among the four 3D classes, all particles mainly clustered into 2 good classes, one of which showed clear density for all protein components of interest whereas the Csy1-Csy2 tail of the other class was largely invisible in the density map (Supplementary information, Figure S5). The full-component class, ∼54 000 particles, was subjected to 3D refinement and yielded a reconstruction of 5.3 Å resolution. Besides the two main classes, one of the minor classes also showed good features in the Csy3-AcrF1 backbone region. Therefore, a total of ∼150 000 particles from these 3 classes were pooled and refined with the rigid backbone masked, which resulted in a 4.5 Å resolution map. After particle polishing, the resolution was improved to 4.2 Å (Supplementary information, Figures S4C and S5), in which the β-strands were clearly separated and bulky amino-acid side chains could be recognized. The local resolutions of all maps were estimated using RESMAP37 (Supplementary information, Figure S6).
Atomic model building
The 3.8 Å map of AcrF1/2-Csy32nt complex backbone showed clear features of amino-acid side chains, but difficult to recognize the topology of main chains. However, the 4.2 Å map of AcrF1/2-Csy20nt complex backbone provided good connectivity information at some areas that were ambiguous in the previous map. The crRNA chain was confidentially identified and modeled, and two copies of AcrF1 atomic model (PDB: 2LW5) were perfectly fitted into the density map and adjusted manually in COOT38, which showed clear side chain densities (Supplementary information, Figure S2E). Locating the crRNA and AcrF1 chains greatly helped the recognition of Csy3 subunit densities. Combining the information of the two maps, we traced the main chain of Csy3 with poly-alanine and the bulky amino-acid side chains were recognized to facilitate sequence registration aided by secondary structure predictions (Supplementary information, Figure S2E). Among the total 342 amino-acid residues in Csy3 polypeptide chain, residues 15-327 were modeled with two disordered loops missing at both termini. The final atomic model contains Csy3-crRNA-AcrF1 components. The Csy4 head was not ideally resolved to facilitate model building but good enough for faithful docking of the reported Csy4 and crRNA hairpin atomic model (PDB: 4AL5) by rigid fitting using SITUS39 and CHIMERA40. The Csy1-Csy2-AcrF2 tail was not modeled.
The model was initially refined with MDFF41, followed by several rounds of iterative manual adjustment in COOT38 and real space refinement using PHENIX42 program with secondary structure and geometry restraints applied. The stereochemical parameters of the model were assessed with MOLPROBITY43 in PHENIX42 package. The detailed image processing and model refinement statistics were summarized in Supplementary information, Table S1.
For other reconstructed maps in which AcrF1 molecules bind with different modes, the individual models of Csy3, Csy4, crRNA and AcrF1 were rigidly fitted into the corresponding positions using SITUS39 and CHIMERA40. The resulting complex pseudo-atomic models were refined in real space using PHENIX42 with secondary structure restraints applied.
Structure analysis and visualization
The reconstructed maps and atomic models were visualized using CHIMERA40 and analyzed using the wrapped applications. All EM density figures were rendered by CHIMERA40 and cartoon representations of atomic models were generated with PYMOL44 software.
Data availability
All the cryo-EM maps used in the structural analysis have been deposited in the EMDB database under the accession codes EMD-6729 (AcrF1/2-Csy32nt complex, mode A backbone), EMD-6730 (AcrF1-Csy32nt complex, mode B), EMD-6731 (AcrF1/2-Csy20nt complex, backbone) and EMD-6728 (AcrF1/2-Csy20nt complex, full component), respectively. The Csy3-crRNA-AcrF1 subcomplex atomic models with 32- and 20-nt spacer sequences have been deposited in the PDB database with entry codes 5XLO and 5XLP, respectively.
Author Contributions
RP, YX, PW, JW, NG and GFG conceived and supervised the study. YX and TZ purified the complex samples and conducted biochemical studies on these samples. RP, YX, TZ and PW prepared the cryo-EM samples and collected data. RP, NL, YC and XZ did image processing and reconstruction. JQ, JW and RP built the atomic model. RP, JQ, NL and YS analyzed the structure. XZ and NG helped map interpretation and structural analysis. RP, YX, TZ and YS prepared the manuscript draft. MW, JW, NG and GFG revised the manuscript and provided intensive discussions. All authors participated in the discussion and manuscript editing.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank all staffs at the National Center for Protein Science Shanghai (NCPSS) cryo-EM department and Xiaojun Huang, Gang Ji and other staffs at the Center of Biological Imaging (IBP, Chinese Academy of Sciences (CAS)) for assistance with data collection. We are grateful to Dr Guopeng Wang (The Core Facilities at School of Life Sciences, Peking University) and Dr Tie Yang (The State Key Laboratory of Membrane Biology, Institute of Zoology, CAS, Beijing) for technical support in electron microscope operation. This study was supported by the Strategic Priority Research Program of CAS (XDB08020100), the Ministry of Science and Technology of China (973 Project; 2013CB531502 and 2014CB542503). YS is supported by the Excellent Young Scientist Program from the National Natural Science Foundation of China (NSFC; 81622031), the Excellent Young Scientist Program of CAS and the Youth Innovation Promotion Association CAS (2015078). GFG is supported partly as a leading principal investigator of the NSFC Innovative Research Group (81621091).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Size exclusion chromatogram and SDS-PAGE profiles of AcrF1/2-Csy32nt complex and AcrF1/2-Csy20nt complex.
Cryo-EM analysis of AcrF1/2-Csy32nt complex.
Major image processing workflow of AcrF1/2-Csy32nt complex.
Cryo-EM analysis of AcrF1/2-Csy20nt complex.
Major image processing workflow of AcrF1/2-Csy20nt complex.
Local resolution distribution patterns of the four reconstructions used in structural analysis.
Superposition of Csy3 backbone densities of AcrF1/2-Csy complex reconstructions and the self-assembly of Csy3 subunits.
Close up views of AcrF1 binding interfaces at different binding modes.
Statistics of the structural models of Csy3-crRNA32nt-AcrF1 and Csy3-crRNA20nt-AcrF1 sub-complexes refined against the 3.8 Å and 4.2 Å resolution cryo-EM maps.
References
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol 2010; 8:317–327. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- Jore MM, Brouns SJ, van der Oost J. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol 2012 Jun 1. doi:10.1101/cshperspect.a003657 [DOI] [PMC free article] [PubMed]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2013; 493:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 2014; 5:e00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL. Phages fight back: inactivation of the CRISPR-Cas bacterial immune system by anti-CRISPR proteins. PLoS Pathog 2016; 12:e1005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K, Chattopadhyay A, Pratap D. Anti-CRISPR proteins: counterattack of phages on bacterial defense (CRISPR/Cas) system. J Cell Physiol 2017 Mar 1. doi:10.1002/jcp.25877 [DOI] [PubMed]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 2010; 11:181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One 2012; 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 2012; 40:5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li J, Zhao H, et al. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 2015; 163:840–853. [DOI] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol 2011; 18:688–692. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 2011; 18:680–687. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 2012; 46:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Nam K H, Ding F, et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol 2014; 21:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol 2015; 30:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 2014; 345:1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 2014; 515:147–150. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 2015; 526:136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma J, Cheng Z, et al. A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses. Cell Res 2016; 26:1165–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yao D, Xu JG, et al. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat Struct Mol Biol 2016; 23:868–870. [DOI] [PubMed] [Google Scholar]
- Maxwell KL, Garcia B, Bondy-Denomy J, Bona D, Hidalgo-Reyes Y, Davidson AR. The solution structure of an anti-CRISPR protein. Nat Commun 2016; 7:13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Carter J, Rollins MF, et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 2017; 169:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DW, Zhu Y, Staals RH, et al. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 2015; 348:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011; 477:486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 2011; 18:529–536. [DOI] [PubMed] [Google Scholar]
- Jun JW, Kim HJ, Yun SK, Chai JY, Park SC. Eating oysters without risk of vibriosis: application of a bacteriophage against Vibrio parahaemolyticus in oysters. Int J Food Microbiol 2014; 188:31–35. [DOI] [PubMed] [Google Scholar]
- Jun JW, Shin TH, Kim JH, et al. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J Infect Dis 2014; 210:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik M, Kiljunen S. Possibilities of bacteriophage therapy. Duodecim 2016; 132:712–719. [PubMed] [Google Scholar]
- Xu Y, Liu Y, Liu Y, Pei J, Yao S, Cheng C. Bacteriophage therapy against Enterobacteriaceae. Virol Sin 2015; 30:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J Struct Biol 2015; 192:204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013; 10:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 2016 Nov 15. doi:10.7554/eLife.18722 [DOI] [PMC free article] [PubMed]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 1999; 125:185–195. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 2009; 49:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB. Arendall WB 3rd, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL PyMOL molecular graphics system. 2002. Available from: URL: http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Size exclusion chromatogram and SDS-PAGE profiles of AcrF1/2-Csy32nt complex and AcrF1/2-Csy20nt complex.
Cryo-EM analysis of AcrF1/2-Csy32nt complex.
Major image processing workflow of AcrF1/2-Csy32nt complex.
Cryo-EM analysis of AcrF1/2-Csy20nt complex.
Major image processing workflow of AcrF1/2-Csy20nt complex.
Local resolution distribution patterns of the four reconstructions used in structural analysis.
Superposition of Csy3 backbone densities of AcrF1/2-Csy complex reconstructions and the self-assembly of Csy3 subunits.
Close up views of AcrF1 binding interfaces at different binding modes.
Statistics of the structural models of Csy3-crRNA32nt-AcrF1 and Csy3-crRNA20nt-AcrF1 sub-complexes refined against the 3.8 Å and 4.2 Å resolution cryo-EM maps.
Data Availability Statement
All the cryo-EM maps used in the structural analysis have been deposited in the EMDB database under the accession codes EMD-6729 (AcrF1/2-Csy32nt complex, mode A backbone), EMD-6730 (AcrF1-Csy32nt complex, mode B), EMD-6731 (AcrF1/2-Csy20nt complex, backbone) and EMD-6728 (AcrF1/2-Csy20nt complex, full component), respectively. The Csy3-crRNA-AcrF1 subcomplex atomic models with 32- and 20-nt spacer sequences have been deposited in the PDB database with entry codes 5XLO and 5XLP, respectively.