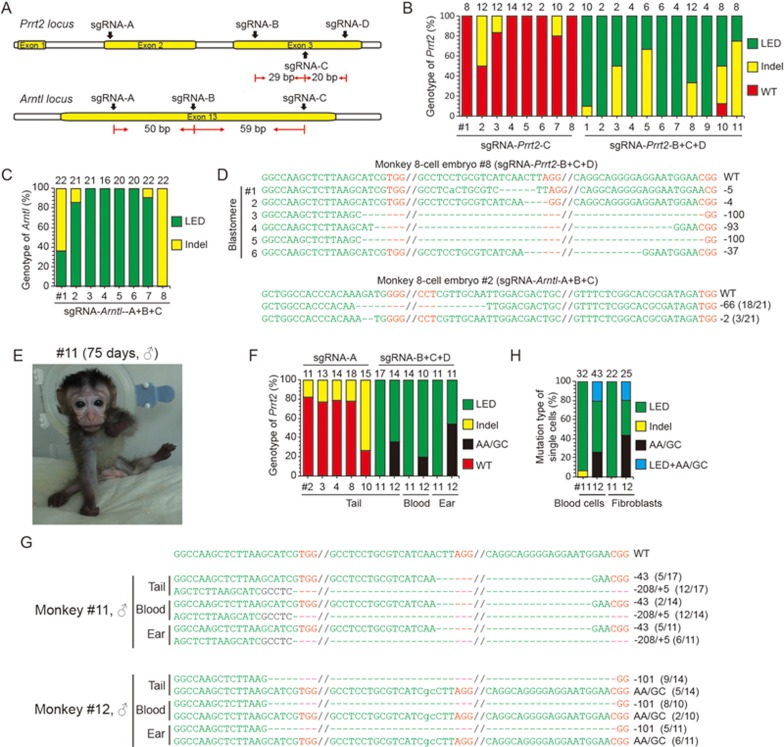
Figure 5.
One-step generation of complete knockout monkey by C-CRISPR. (A) Schematic diagram of sgRNA-targeting sites at Prrt2 and Arntl loci in monkey. (B and C) Percentages of different mutation types for 8-cell embryos with Prrt2 (B) and Arntl (C) targeting. LED, large-fragment exon deletion. Indels, insertion, or deletion of bases. Number above each column, total alleles analyzed from blastomeres of 8-cell embryos with Prrt2 targeting or total TA clones sequenced from 8-cell embryos with Arntl targeting. (D) Genotyping analysis on single blastomeres of 8-cell embryos targeted with sgRNA-Prrt2-B+C+D or whole 8-cell embryos targeted with sgRNA-Arntl-A+B+C. The sgRNA targeted sequences are labeled in green and the PAM sequences are labeled in red; deleted nucleotides are indicated by hyphens. Dashed lines mark the region omitted for clarity. (E) Photograph of Prrt2-knockout monkey (#11). (F) Percentages of different mutation types found in tail, ear, and blood cells from aborted and live monkeys by Prrt2 targeting. Aborted monkeys with sgRNA-Prrt2-A targeting: monkey #2, #3; live monkeys with sgRNA-Prrt2-A targeting: monkey #4, #8, #10; live monkeys with sgRNA-Prrt2-B+C+D targeting: monkey #11, #12. AA/GC, a point mutation in Prrt2(335N to 335A). (G) Representative sequences from ear, tail, and blood cells of monkey #11 and #12 with sgRNA-Prrt2-B+C+D targeting. The sgRNA-targeting sequences are labeled in green and PAM sequences are labeled in red; deleted nucleotides are indicated by hyphens. Dashed lines mark the region omitted for clarity. (H) Single-cell analysis on blood cells and fibroblasts from two live monkeys with Prrt2 editing (#11, #12). Number, total number of cells analyzed.